Anti-Inflammatory Effects of (9Z,11E)-13-Oxooctadeca-9,11-dienoic Acid (13-KODE) Derived from Salicornia herbacea L. on Lipopolysaccharide-Stimulated Murine Macrophage via NF-kB and MAPK Inhibition and Nrf2/HO-1 Signaling Activation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Source
2.3. Purification of (9Z,11E)-13-Oxooctadeca-9,11-dienoic Acid (13-KODE)
2.4. Structural Analysis of the Isolated Sample
2.5. Antioxidant Assay
2.6. Cell Line and Culture Conditions
2.7. Cell Proliferation Assay
2.8. NO Assay
2.9. Quantitative Real Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.10. Enzyme-Linked Immunosorbent Assay (ELISA) for Cytokines
2.11. Western Blot Analysis
2.12. Immunofluorescence Staining
2.13. Determination of Cellular ROS by Invitrogen® CellROX® Green Reagent
2.14. Statistical Analysis
3. Results
3.1. Purification of an Antioxidant Derived from S. herbacea L.
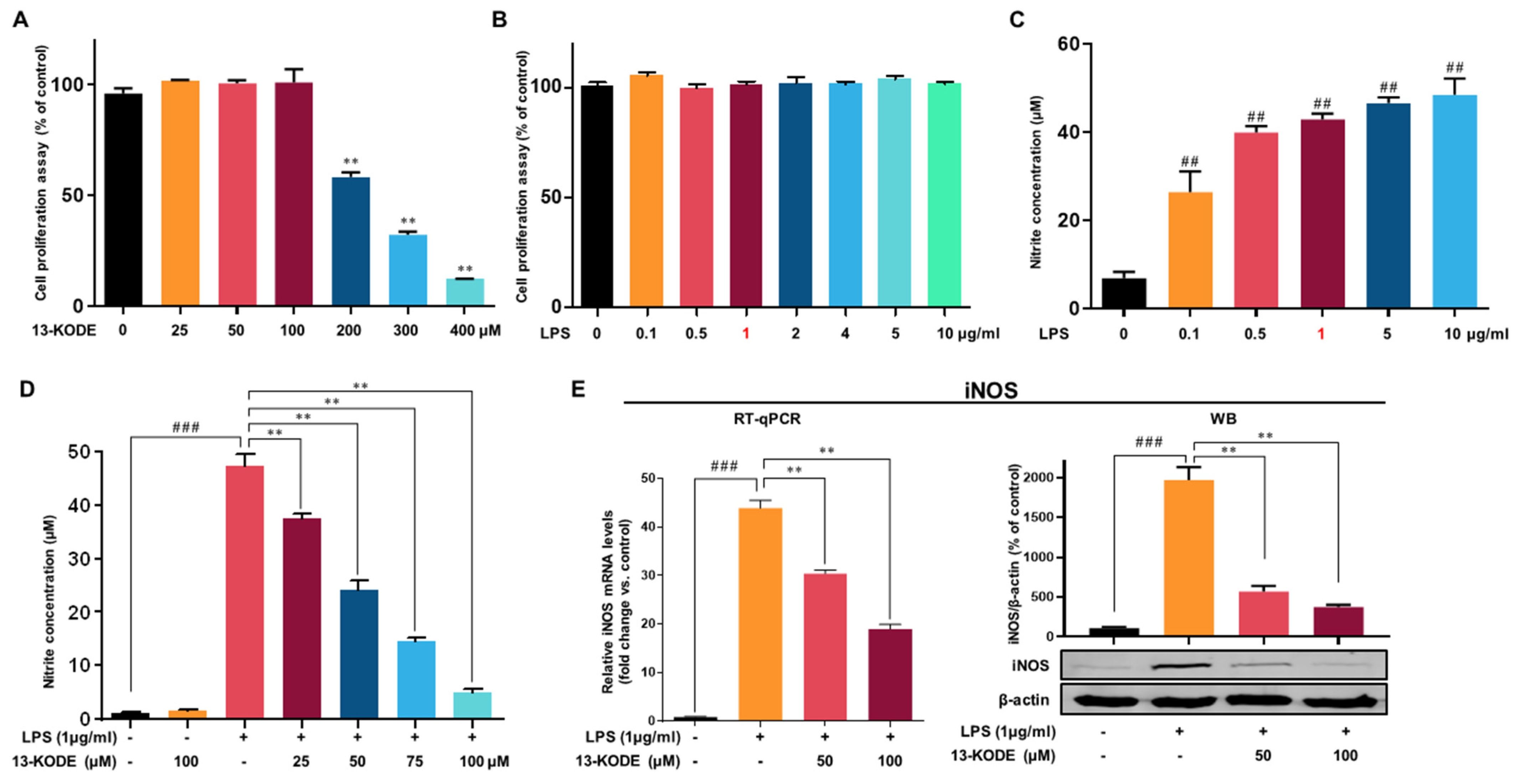
3.2. Structure of Isolated Antioxidant and the Effect of 13-KODE on Cell Proliferation
3.3. Effect of 13-KODE on LPS-Stimulated Production of Proinflammatory Mediators in RAW 264.7 Cells
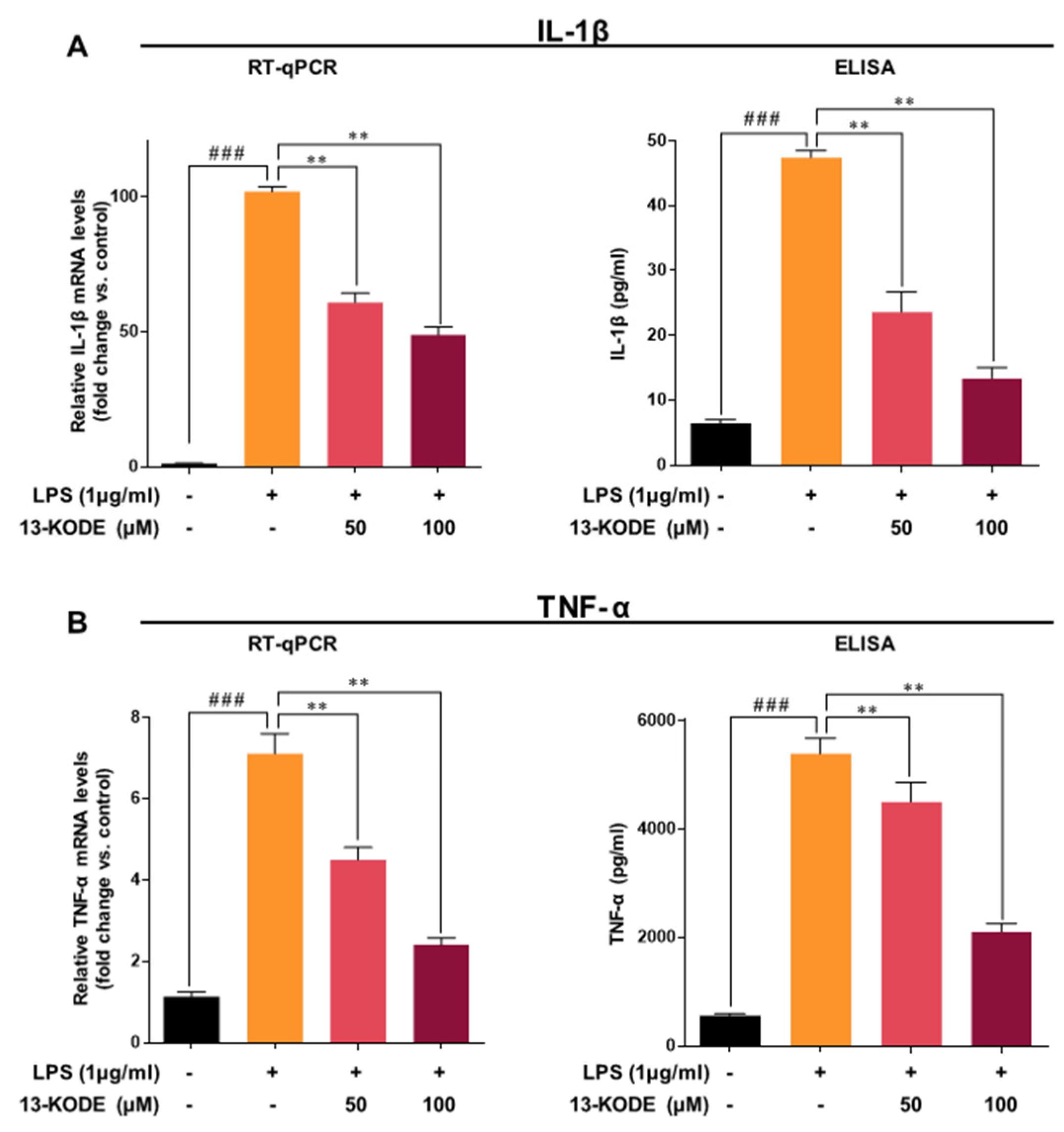
3.4. 13-KODE Inhibits LPS-Stimulated Proinflammatory Cytokines in RAW 264.7 Macrophage Cells
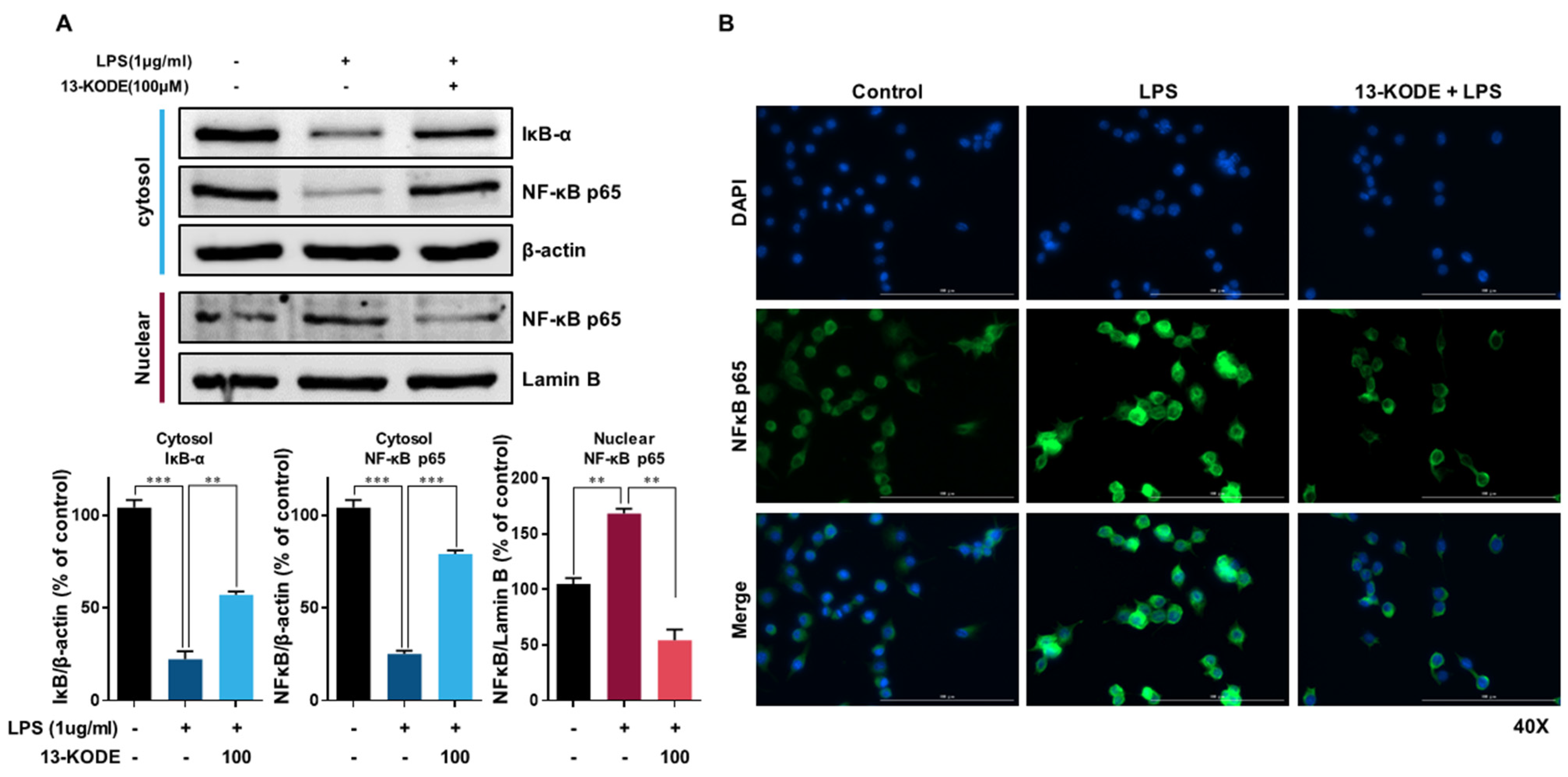
3.5. The Effect of 13-KODE on NF-κB Signaling
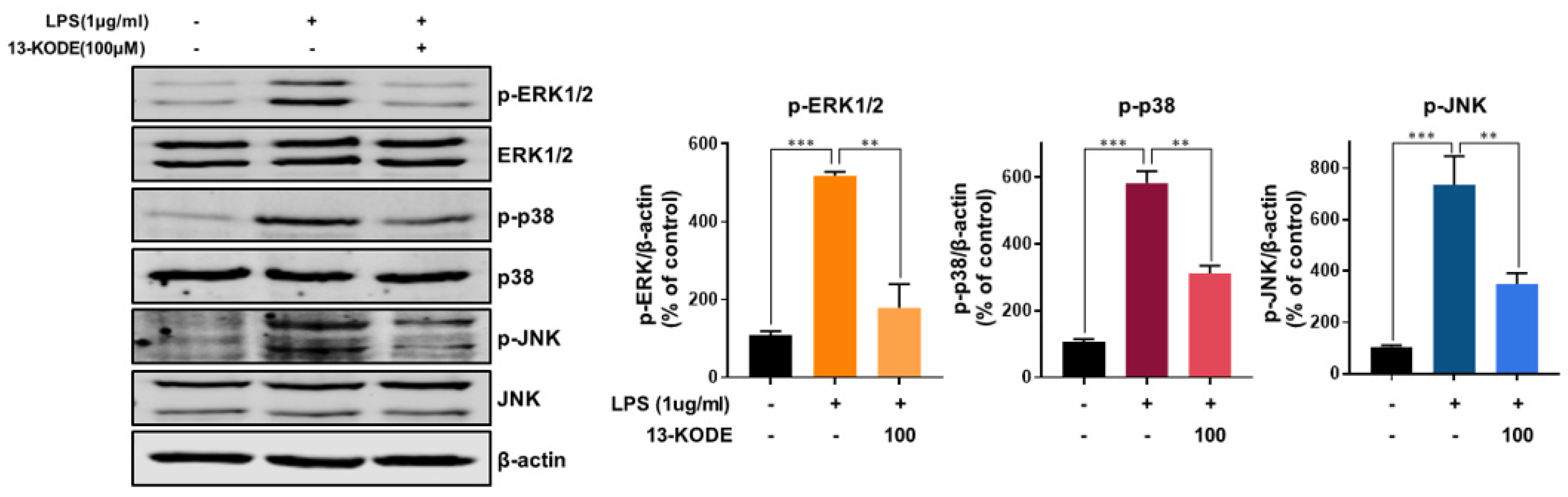
3.6. 13-KODE Inhibits LPS-Induced MAPK Activation in RAW 264.7 Macrophages
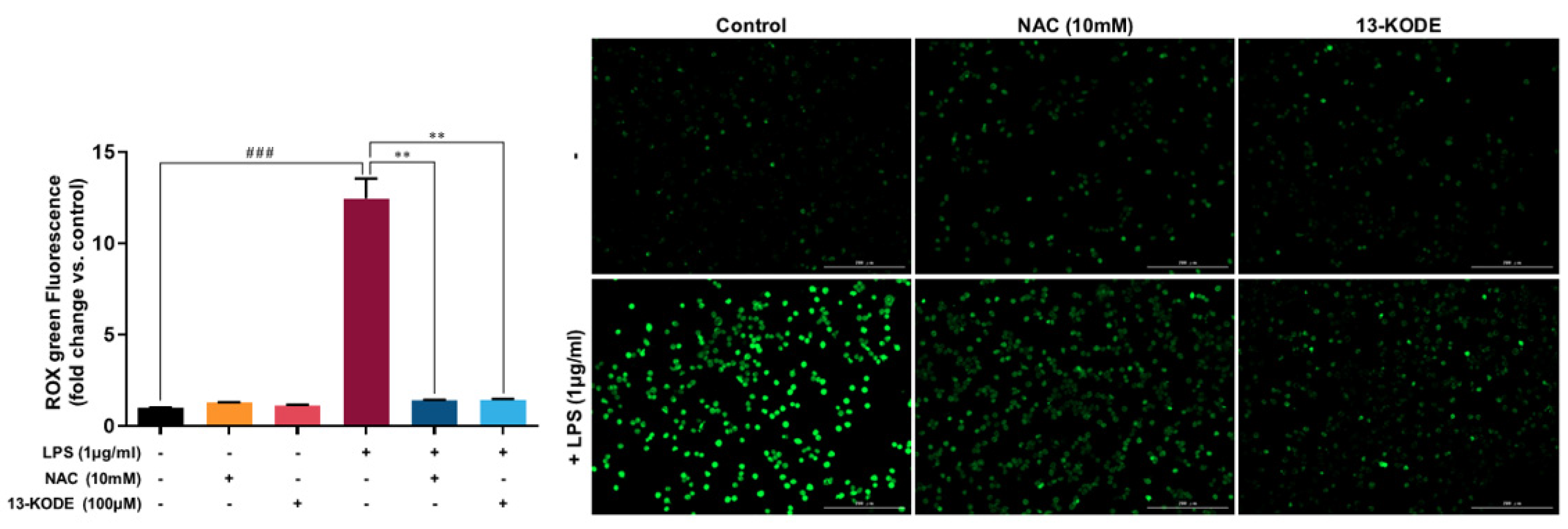
3.7. 13-KODE Reduced LPS-Induced ROS Accumulation in RAW 264.7 Cells
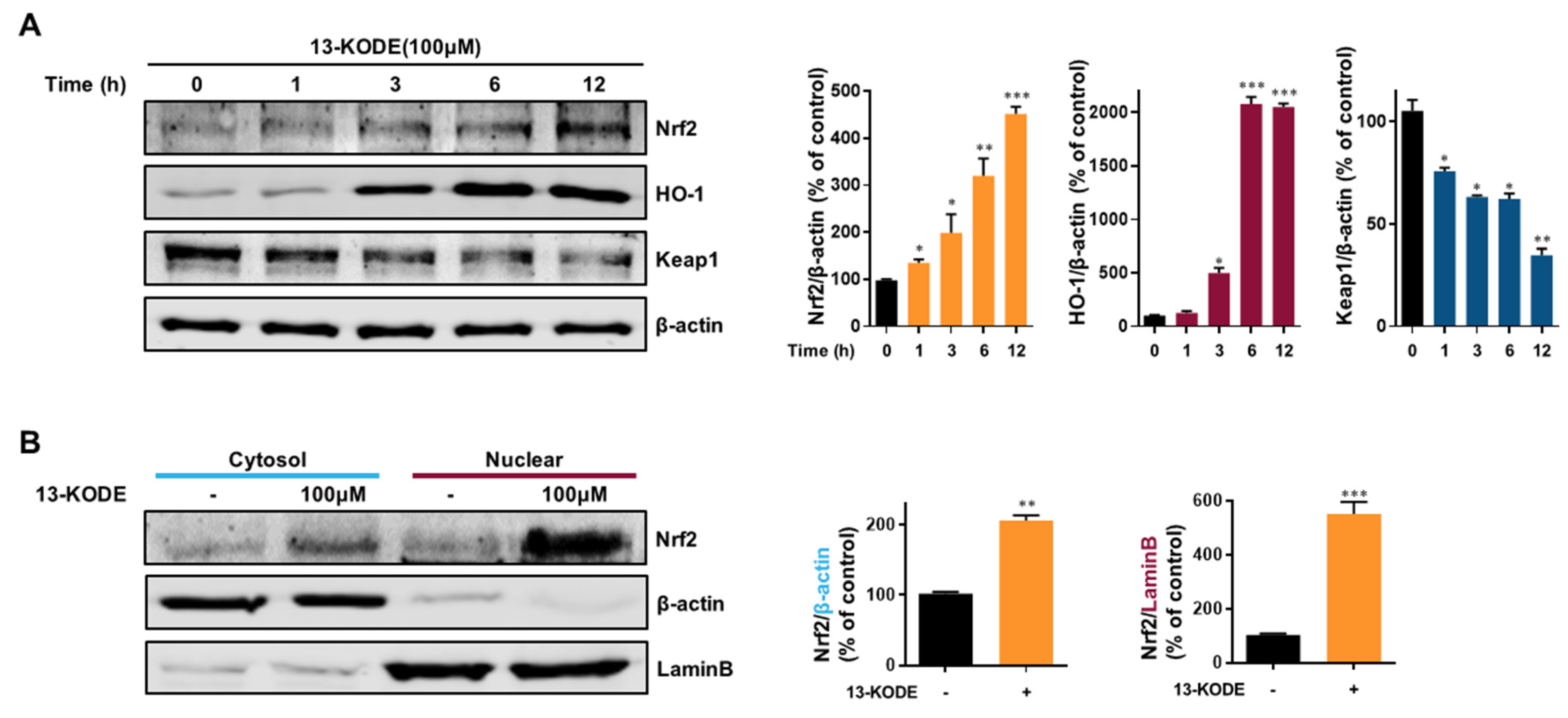
3.8. Induction of HO-1 and Nfe2I2 Expression by 13-KODE in RAW 264.7 Cells
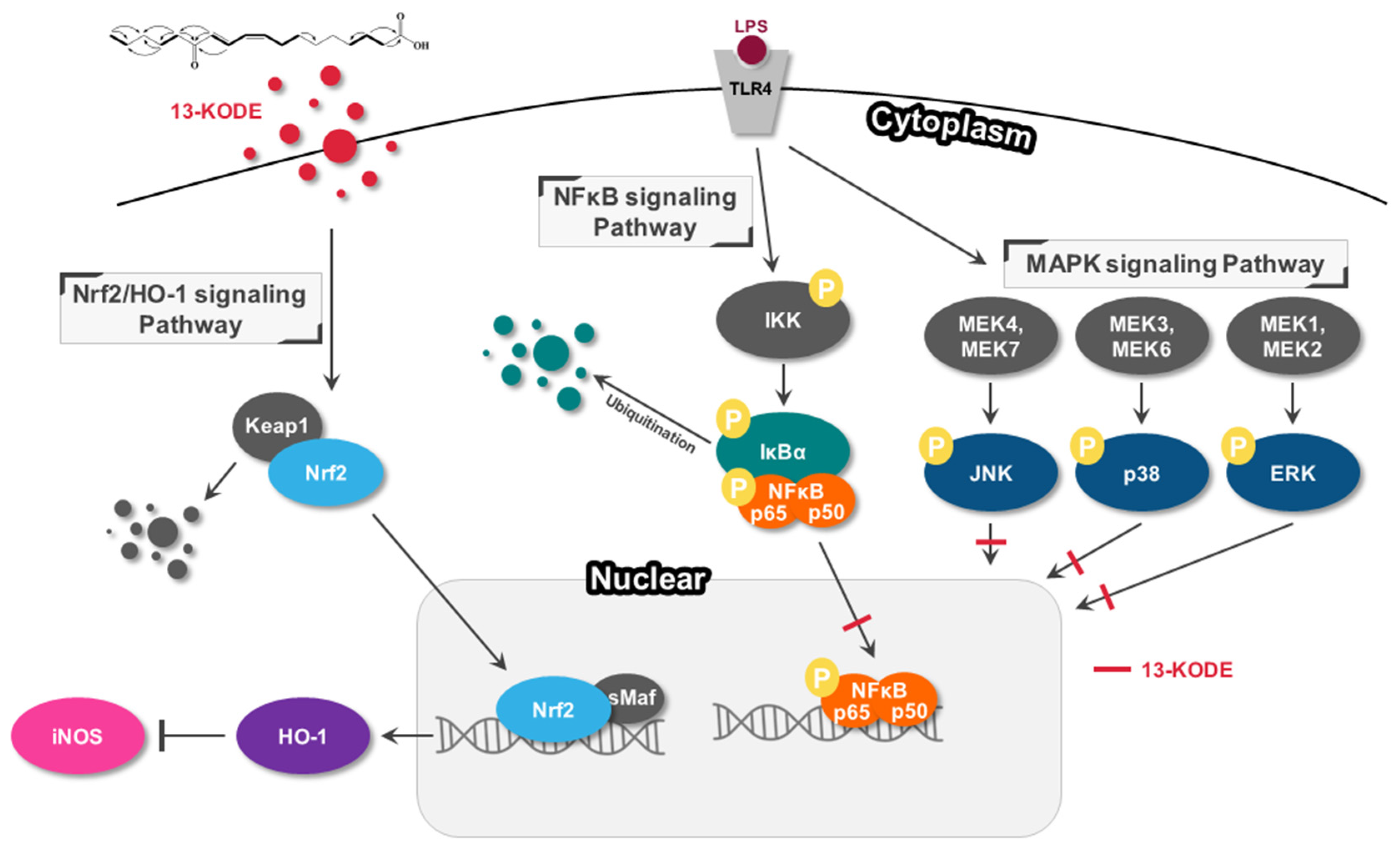
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrero-Miliani, L.; Nielsen, O.H.; Andersen, P.S.; Girardin, S.E. Chronic Inflammation: Importance of NOD2 and NALP3 in Interleukin-1beta Generation. Clin. Exp. Immunol. 2007, 147, 227–235. [Google Scholar] [CrossRef]
- Rock, K.L.; Kono, H. The Inflammatory Response to Cell Death. Annu. Rev. Pathol. 2008, 3, 99–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuschieri, J.; Maier, R.V. Oxidative Stress, Lipid Rafts, and Macrophage Reprogramming. Antioxid. Redox Signal. 2007, 9, 1485–1497. [Google Scholar] [CrossRef]
- Lucas, K.; Maes, M. Role of the Toll Like Receptor (TLR) Radical Cycle in Chronic Inflammation: Possible Treatments Targeting the TLR4 Pathway. Mol. Neurobiol. 2013, 48, 190–204. [Google Scholar] [CrossRef]
- Leiro, J.; Alvarez, E.; Arranz, J.A.; Laguna, R.; Uriarte, E.; Orallo, F. Effects of Cis-Resveratrol on Inflammatory Murine Macrophages: Antioxidant Activity and Down-Regulation of Inflammatory Genes. J. Leukoc. Biol. 2004, 75, 1156–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.W.; Chiou, W.F.; Chao, S.H.; Lee, M.H.; Chen, C.C.; Tsai, Y.C. Ligustilide Prevents LPS-induced iNOS Expression in RAW 264.7 Macrophages by Preventing ROS Production and Down-Regulating the MAPK, NF-kappaB and AP-1 Signaling Pathways. Int. Immunopharmacol. 2011, 11, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Ohigashi, H. Targeting NOX, INOS and COX-2 in Inflammatory Cells: Chemoprevention Using Food Phytochemicals. Int. J. Cancer 2007, 121, 2357–2363. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.K.; Albrecht, F.; Rohrer, C.; Koeberle, A.; Werz, O.; Schlormann, W.; Glei, M.; Lorkowski, S.; Wallert, M. Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2. Nutrients 2021, 13, 4437. [Google Scholar] [CrossRef] [PubMed]
- Klapproth, J.M.; Sasaki, M. Bacterial Induction of Proinflammatory Cytokines in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2010, 16, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Shi, R.; Wang, S.; Liu, Q.; Zhang, H.; Chen, X. Intermedin Inhibits the Ox-LDL-Induced Inflammation in RAW264.7 Cells by Affecting Fatty Acid-Binding Protein 4 through the PKA Pathway. Front. Pharmacol. 2021, 12, 724777. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Mackman, N. LPS Induction of Gene Expression in Human Monocytes. Cell Signal. 2001, 13, 85–94. [Google Scholar] [CrossRef]
- Huang, B.P.; Lin, C.H.; Chen, H.M.; Lin, J.T.; Cheng, Y.F.; Kao, S.H. AMPK Activation Inhibits Expression of Proinflammatory Mediators through Downregulation of PI3K/p38 MAPK and NF-kappaB Signaling in Murine Macrophages. DNA Cell Biol. 2015, 34, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liao, F.; Luo, G.; Qian, Y.; He, X.; Xu, W.; Ding, S.; Pu, J. NR1D1 Deletion Induces Rupture-Prone Vulnerable Plaques by Regulating Macrophage Pyroptosis via the NF-kappaB/NLRP3 Inflammasome Pathway. Oxid. Med. Cell. Longev. 2021, 5217572. [Google Scholar]
- Huang, P.; Hong, J.; Mi, J.; Sun, B.; Zhang, J.; Li, C.; Yang, W. Polyphenols Extracted from Enteromorpha Clathrata Alleviates Inflammation in Lipopolysaccharide-Induced RAW 264.7 Cells by Inhibiting the MAPKs/NF-kappaB Signaling Pathways. J. Ethnopharmacol. 2021, 286, 114897. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-Activated Protein (MAP) Kinase Pathways: Regulation and Physiological Functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- Huang, Y.; Li, W.; Su, Z.Y.; Kong, A.N. The Complexity of the Nrf2 Pathway: Beyond the Antioxidant Response. J. Nutr. Biochem. 2015, 26, 1401–1413. [Google Scholar] [CrossRef]
- Nakajima, S.; Kitamura, M. Bidirectional Regulation of NF-kappaB by Reactive Oxygen Species: A Role of Unfolded Protein Response. Free Radic. Biol. Med. 2013, 65, 162–174. [Google Scholar] [CrossRef]
- Fusco, R.; Cordaro, M.; Siracusa, R.; Peritore, A.F.; D’Amico, R.; Licata, P.; Crupi, R.; Gugliandolo, E. Effects of Hydroxytyrosol against Lipopolysaccharide-Induced Inflammation and Oxidative Stress in Bovine Mammary Epithelial Cells: A Natural Therapeutic Tool for Bovine Mastitis. Antioxidants. 2020, 9, 693. [Google Scholar] [CrossRef] [PubMed]
- Crupi, R.; Palma, E.; Siracusa, R.; Fusco, R.; Gugliandolo, E.; Cordaro, M.; Impellizzeri, D.; De Caro, C.; Calzetta, L.; Cuzzocrea, S.; et al. Protective Effect of Hydroxytyrosol against Oxidative Stress Induced by the Ochratoxin in Kidney Cells: In vitro and in vivo Study. Front. Vet. Sci. 2020, 7, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Yuan, Y. Accelerated Inflammation and Oxidative Stress Induced by LPS in Acute Lung Injury: Iotanhibition by ST1926. Int. J. Mol. Med. 2018, 41, 3405–3421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meital, L.T.; Windsor, M.T.; Perissiou, M.; Schulze, K.; Magee, R.; Kuballa, A.; Golledge, J.; Bailey, T.G.; Askew, C.D.; Russell, F.D. Omega-3 Fatty Acids Decrease Oxidative Stress and Inflammation in Macrophages from Patients with Small Abdominal Aortic Aneurysm. Sci. Rep. 2019, 9, 12978. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, R.; Fusco, R.; Peritore, A.F.; Cordaro, M.; D’Amico, R.; Genovese, T.; Gugliandolo, E.; Crupi, R.; Smeriglio, A.; Mandalari, G.; et al. The Antioxidant and Anti-Inflammatory Properties of Anacardium occidentale L. Cashew Nuts in a Mouse Model of Colitis. Nutrients 2020, 12, 834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xu, X.; Liu, Z.; Sun-Waterhouse, D.; Wang, J.; Ma, C.; Waterhouse, G.I.N.; Kang, W. Effects of Edpetiline from Fritillaria on Inflammation and Oxidative Stress induced by LPS Stimulation in RAW264.7 Macrophages. Acta Biochim. Biophys. Sin. 2021, 53, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, Y.; Bae, M.; Park, Y.K.; Lee, J.Y. Astaxanthin Inhibits Alcohol-Induced Inflammation and Oxidative Stress in Macrophages in a Sirtuin 1-Dependent Manner. J. Nutr. Biochem. 2020, 85, 108477. [Google Scholar] [CrossRef] [PubMed]
- Deshane, J.; Wright, M.; Agarwal, A. Heme Oxygenase-1 Expression in Disease States. Acta Biochim. Pol. 2005, 52, 273–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 System in Development, Oxidative Stress Response and Diseases: An Evolutionarily Conserved Mechanism. Cell Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.A.; Hyun, J.W. Oxidative Stress, Nrf2, and Epigenetic Modification Contribute to Anticancer Drug Resistance. Toxicol. Res. 2017, 33, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Seo, J.Y.; Oh, J.; Jang, Y.K.; Lee, C.H.; Kim, J.S. Neuroprotective Effect of Halophyte Salicornia herbacea L. Is Mediated by Activation of Heme Oxygenase-1 in Mouse Hippocampal HT22 Cells. J. Med. Food 2017, 20, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Ko, S.K.; Choi, J.G.; Chung, S.H. Salicornia Herbacea Prevents High Fat Diet-Induced Hyperglycemia and Hyperlipidemia in ICR Mice. Arch. Pharm. Res. 2006, 29, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Park, S.H.; Seo, D.H.; Lee, J.H.; Hong, S.W.; Hong, S.S. Antioxidative and Skin-Whitening Effect of an Aqueous Extract of Salicornia Herbacea. Biosci. Biotechnol. Biochem. 2009, 73, 552–556. [Google Scholar] [CrossRef] [Green Version]
- Shen, T.; Heo, S.I.; Wang, M.H. Involvement of the p38 MAPK and ERK Signaling Pathway in the Anti-Melanogenic Effect of Methyl 3,5-dicaffeoyl Quinate in B16F10 Mouse Melanoma Cells. Chem. Biol. Interact. 2012, 199, 106–111. [Google Scholar] [CrossRef]
- Takahashi, H.; Kim, Y.I.; Hirai, S.; Goto, T.; Ohyane, C.; Tsugane, T.; Konishi, C.; Fujii, T.; Inai, S.; Iijima, Y.; et al. Comparative and Stability Analyses of 9- and 13-Oxo-octadecadienoic Acids in Various Species of Tomato. Biosci. Biotechnol. Biochem. 2011, 75, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Vangaveti, V.N.; Jansen, H.; Kennedy, R.L.; Malabu, U.H. Hydroxyoctadecadienoic Acids: Oxidised Derivatives of Linoleic Acid and Their Role in Inflammation Associated with Metabolic Syndrome and Cancer. Eur. J. Pharm. 2016, 785, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free-Radical Method to Evaluate Antioxidant Activity. Food Sci. Technol. -Lebensm. -Wiss. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kim, J.Y.; Lee, Y.G.; Lee, H.J.; Shim, H.J.; Lee, J.H.; Kim, S.J.; Ham, K.S.; Moon, J.H. Four New Dicaffeoylquinic Acid Derivatives from Glasswort (Salicornia herbacea L.) and Their Antioxidative Activity. Molecules 2016, 21, 1097. [Google Scholar] [CrossRef] [Green Version]
- Ha, B.J.; Lee, S.H.; Kim, H.J.; Lee, J.Y. The Role of Salicornia Herbacea in Ovariectomy-Induced Oxidative Stress. Biol. Pharm. Bull. 2006, 29, 1305–1309. [Google Scholar] [CrossRef] [Green Version]
- Im, S.A.; Kim, K.; Lee, C.K. Immunomodulatory Activity of Polysaccharides Isolated from Salicornia Herbacea. Int. Immunopharmacol. 2006, 6, 1451–1458. [Google Scholar] [CrossRef]
- Ko, Y.C.; Choi, H.S.; Kim, J.H.; Kim, S.L.; Yun, B.S.; Lee, D.S. Coriolic Acid (13-(S)-Hydroxy-9Z, 11E-octadecadienoic Acid) from Glasswort (Salicornia herbacea L.) Suppresses Breast Cancer Stem Cell through the Regulation of c-Myc. Molecules 2020, 25, 4950. [Google Scholar] [CrossRef]
- Ryu, D.S.; Kim, S.H.; Lee, D.S. Anti-Proliferative Effect of Polysaccharides from Salicornia Herbacea on Induction of G2/M arrest and Apoptosis in Human Colon Cancer Cells. J. Microbiol. Biotechnol. 2009, 19, 1482–1489. [Google Scholar] [CrossRef] [PubMed]
- Reinaud, O.; Delaforge, M.; Boucher, J.L.; Rocchiccioli, F.; Mansuy, D. Oxidative Metabolism of Linoleic Acid by Human Leukocytes. Biochem. Biophys. Res. Commun. 1989, 161, 883–891. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Sang, Y.; Liu, W.; Yu, W.; Wang, X. Anti-Inflammatory Effects of Gingerol on Lipopolysaccharide-Stimulated RAW 264.7 Cells by Inhibiting NF-kappaB Signaling Pathway. Inflammation 2018, 41, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, T.B.; Moon, K.A.; Kim, T.J.; Shin, D.; Cho, Y.S.; Moon, H.B.; Lee, K.Y. Regulation of Pro-Inflammatory Responses by Lipoxygenases via Intracellular Reactive Oxygen Species in vitro and in vivo. Exp. Mol. Med. 2008, 40, 461–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Jia, Z.; Li, Y.R. Nrf2 Signaling in Macrophages. React. Oxyg. Species (Apex) 2016, 2, 417–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habtemariam, S. The Nrf2/HO-1 Axis as Targets for Flavanones: Neuroprotection by Pinocembrin, Naringenin, and Eriodictyol. Oxid. Med. Cell. Longev. 2019, 2019, 4724920. [Google Scholar] [CrossRef] [PubMed]
- Sauer, L.A.; Dauchy, R.T.; Blask, D.E.; Armstrong, B.J.; Scalici, S. 13-Hydroxyoctadecadienoic Acid Is the Mitogenic Signal for Linoleic Acid-Dependent Growth in Rat Hepatoma 7288CTC in vivo. Cancer Res. 1999, 59, 4688–4692. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, Y.-C.; Choi, H.S.; Kim, S.-L.; Yun, B.-S.; Lee, D.-S. Anti-Inflammatory Effects of (9Z,11E)-13-Oxooctadeca-9,11-dienoic Acid (13-KODE) Derived from Salicornia herbacea L. on Lipopolysaccharide-Stimulated Murine Macrophage via NF-kB and MAPK Inhibition and Nrf2/HO-1 Signaling Activation. Antioxidants 2022, 11, 180. https://doi.org/10.3390/antiox11020180
Ko Y-C, Choi HS, Kim S-L, Yun B-S, Lee D-S. Anti-Inflammatory Effects of (9Z,11E)-13-Oxooctadeca-9,11-dienoic Acid (13-KODE) Derived from Salicornia herbacea L. on Lipopolysaccharide-Stimulated Murine Macrophage via NF-kB and MAPK Inhibition and Nrf2/HO-1 Signaling Activation. Antioxidants. 2022; 11(2):180. https://doi.org/10.3390/antiox11020180
Chicago/Turabian StyleKo, Yu-Chan, Hack Sun Choi, Su-Lim Kim, Bong-Sik Yun, and Dong-Sun Lee. 2022. "Anti-Inflammatory Effects of (9Z,11E)-13-Oxooctadeca-9,11-dienoic Acid (13-KODE) Derived from Salicornia herbacea L. on Lipopolysaccharide-Stimulated Murine Macrophage via NF-kB and MAPK Inhibition and Nrf2/HO-1 Signaling Activation" Antioxidants 11, no. 2: 180. https://doi.org/10.3390/antiox11020180





