New Insights into the Cellular Toxicity of Carbon Quantum Dots to Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Reagents
2.2. Characterization of CQDs
2.3. Bacteriostatic Experiment
2.4. MTT Assay
2.5. Oxidative Stress Detection
2.6. Interaction of CQDs with the Bacterial Membrane
2.7. SEM and TEM Characterizations
2.8. Comet Assay
2.9. Zeta Potential and Osmotic Pressure Measurements
3. Results
3.1. Characterization of CQDs
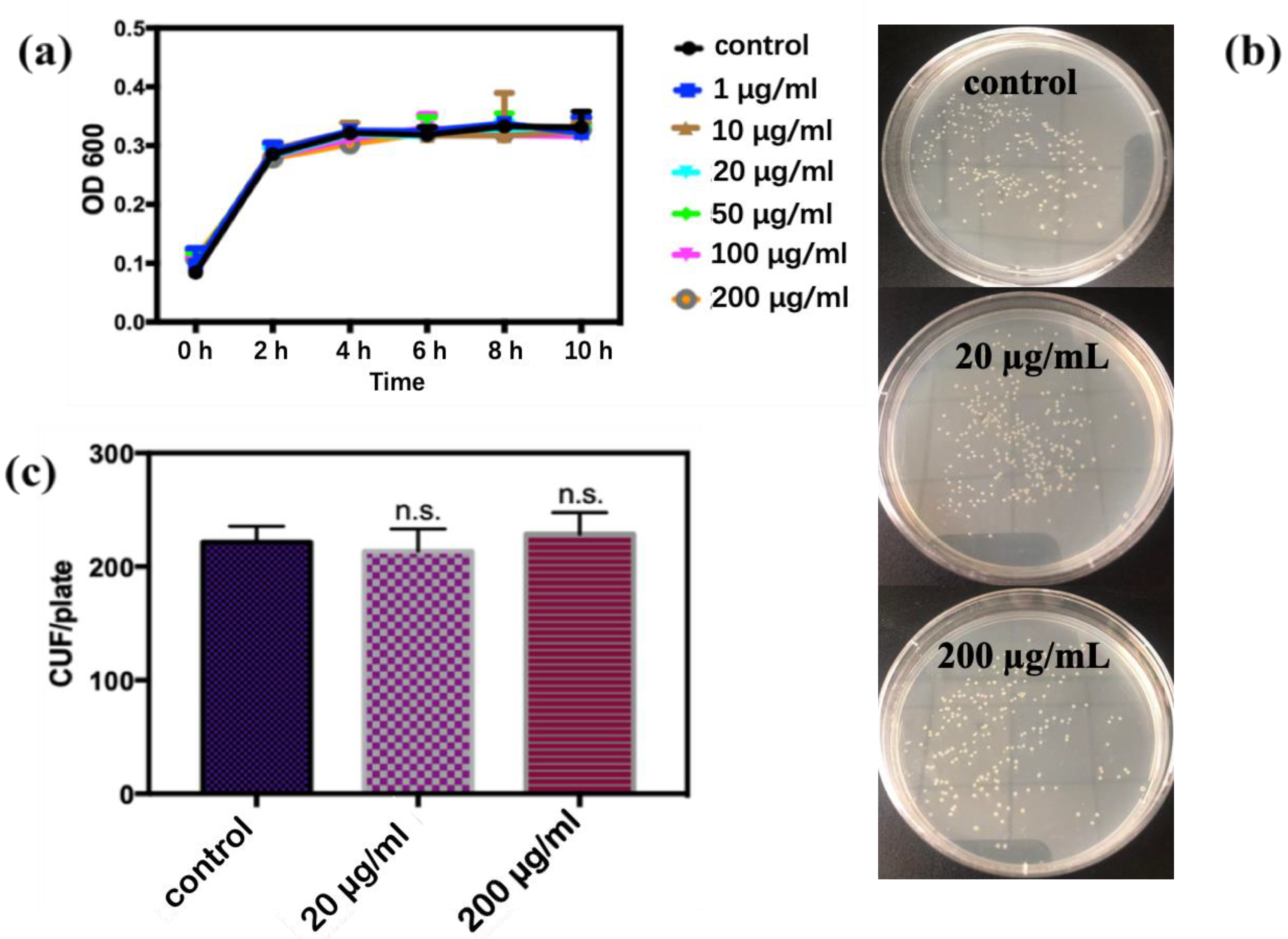
3.2. Bacteriostatic Experiment
3.3. MTT Assay
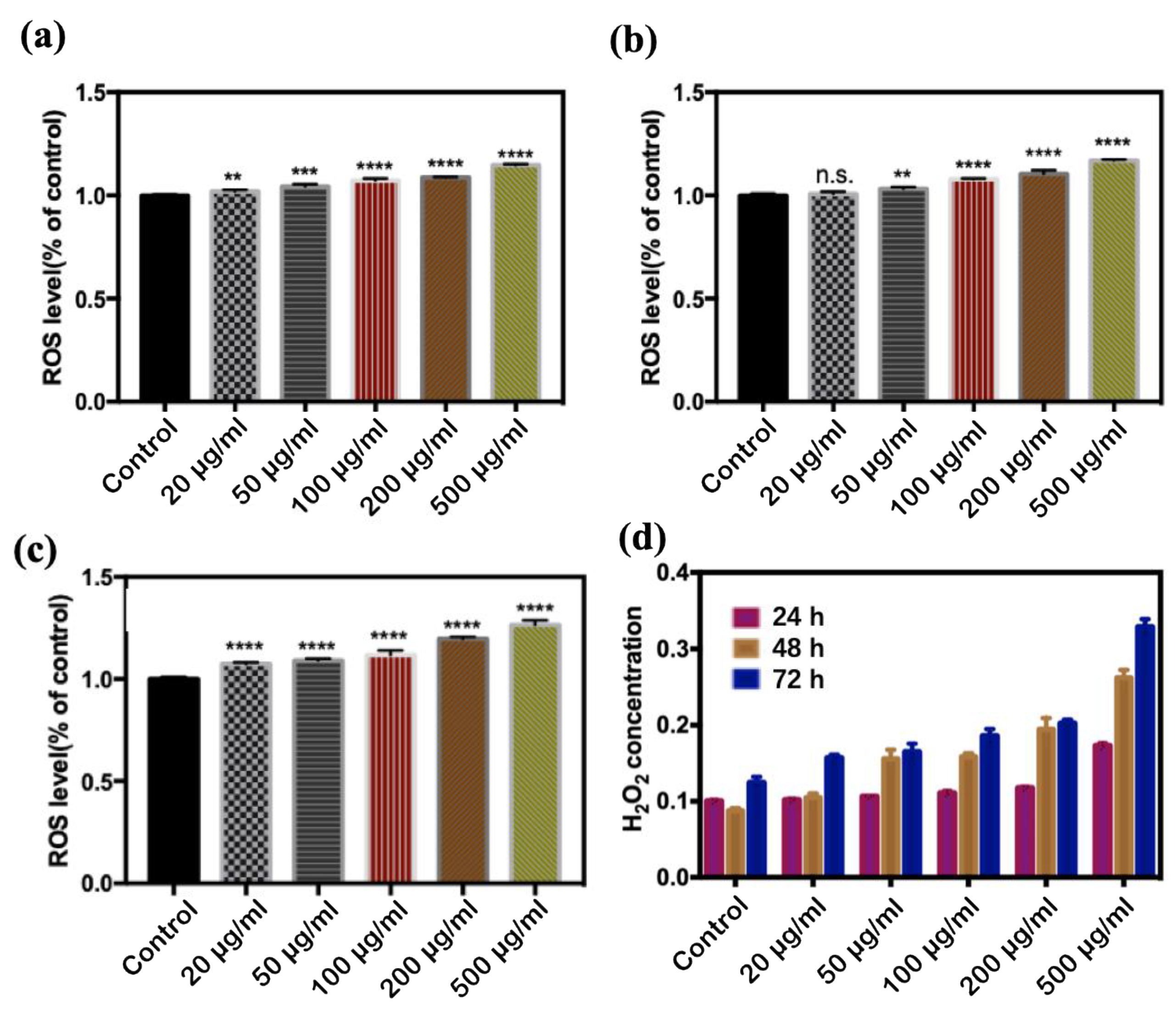
3.4. Oxidative Stress
3.5. Interaction of CQDs with the Bacterial Membrane
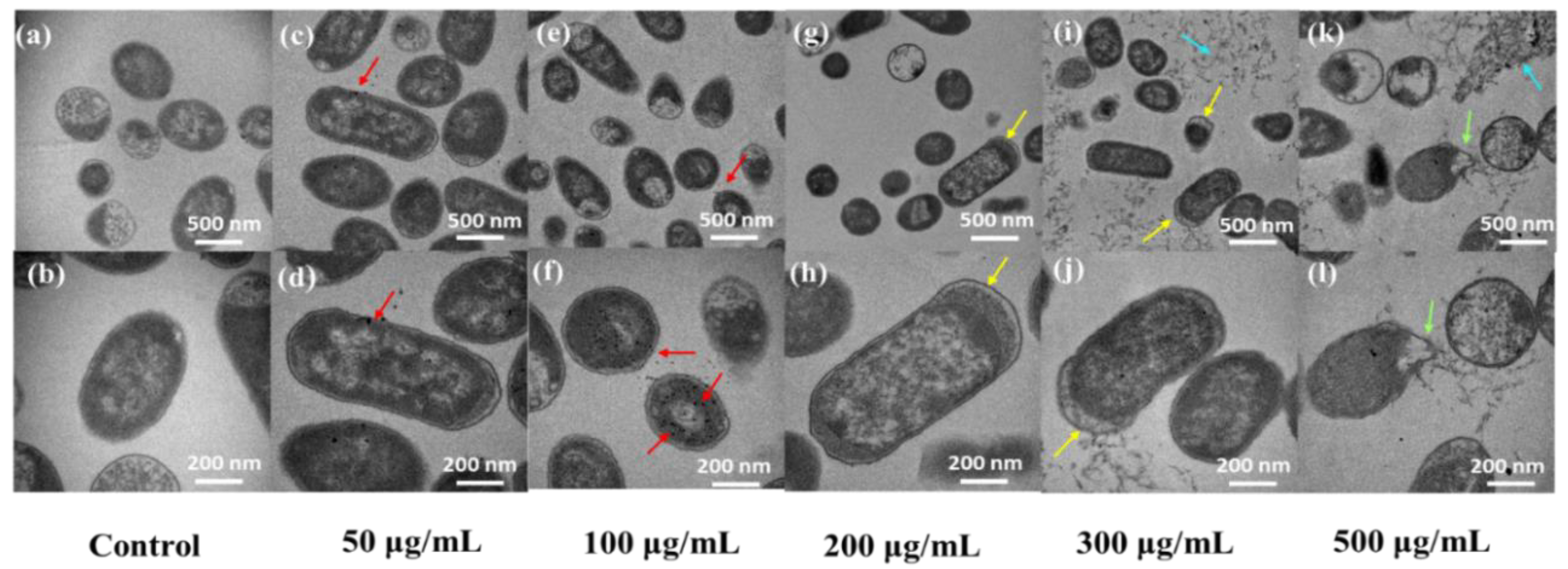
3.6. Characterizations of SEM and TEM on E. coli Exposed to CQDs
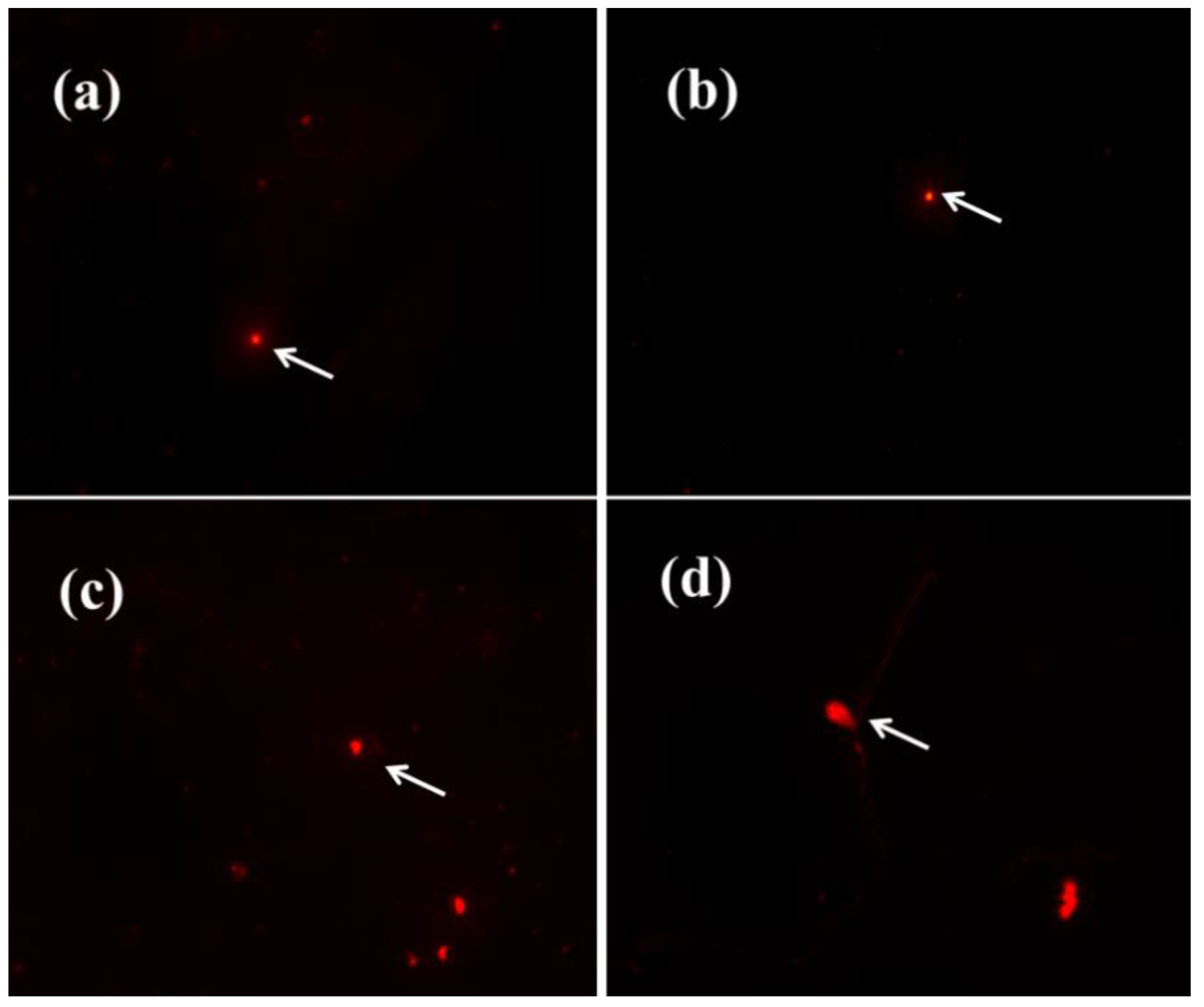
3.7. Comet Assay
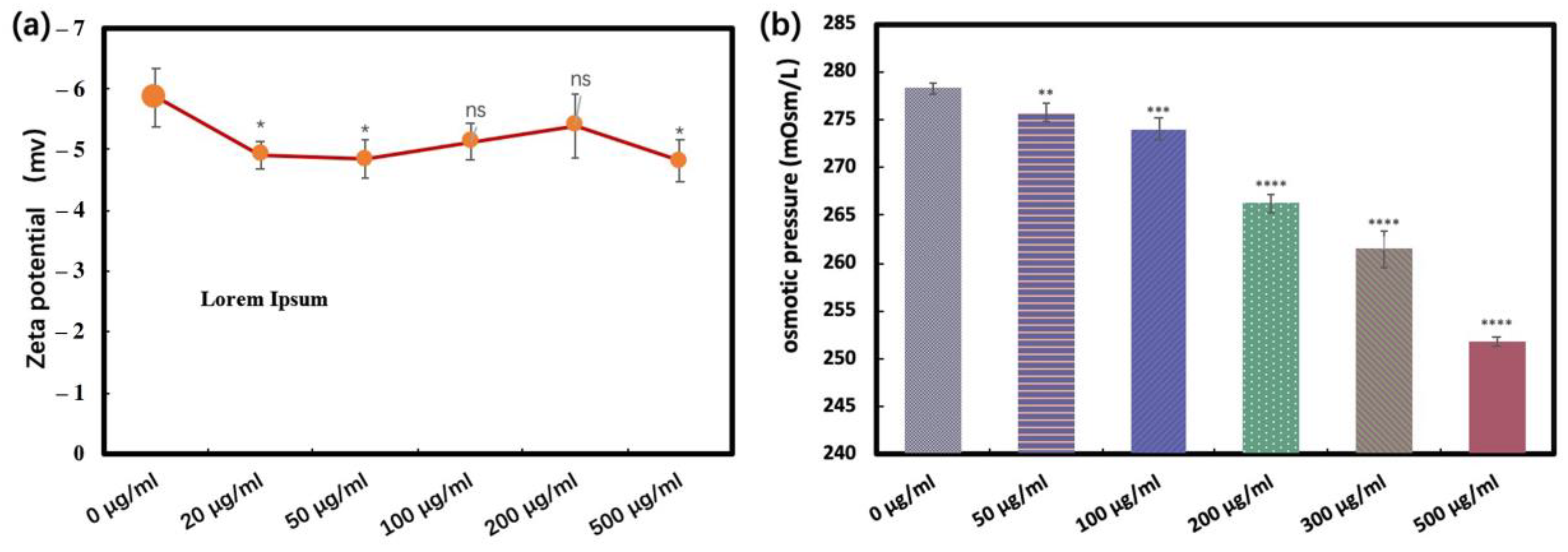
3.8. Zeta Potential and Osmotic Pressure
4. Discussion
4.1. Characterization of CQDs
4.2. Evaluation of Cytotoxicity of CQDs to E. coli
4.2.1. Analysis of Antibacterial Activity after a Short Exposure Time
4.2.2. Survival Rate with the Time of Exposure
4.2.3. Oxidative Stress
4.2.4. Changes in the Bacterial Cytoderm
4.3. Cytotoxic Mechanism of CQDs in E. coli
4.3.1. Microscopic Morphological Analysis
4.3.2. Analysis of DNA Damage
4.3.3. Zeta Potential
4.3.4. Changes in Osmotic Pressure of E. coli
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lim, S.Y.; Shen, W.; Gao, Z. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Molaei, M.J. A review on nanostructured carbon quantum dots and their applications in biotechnology, sensors, and chemiluminescence. Talanta 2019, 196, 456–478. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Jiang, X.; Qin, D.; Mo, G.; Zheng, X.; Deng, B. Facile syntheses of s, n-codoped carbon quantum dots and their applications to a novel off–on nanoprobe for detection of 6-thioguanine and its bioimaging. ACS Sustain. Chem. Eng. 2019, 7, 16112–16120. [Google Scholar] [CrossRef]
- Xiao, A.; Wang, C.; Chen, J.; Guo, R.; Yan, Z.; Chen, J. Carbon and metal quantum dots toxicity on the microalgae Chlorella pyrenoidosa. Ecotoxicol. Environ. Saf. 2016, 133, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Qu, J.; Yao, Y.; Zhang, Y.; Ma, Y.; Wu, D.; Cao, Y.; Yang, M.; Zhang, Y.; Tang, M. N-doped carbon dots triggered the induction of ROS-mediated cytoprotective autophagy in Hepa1-6 cells. Chemosphere 2020, 251, 126440. [Google Scholar] [CrossRef]
- Yan, J.; Hou, S.; Yu, Y.; Qiao, Y.; Xiao, T.; Mei, Y.; Zhang, Z.; Wang, B.; Huang, C.C.; Lin, C.H. The effect of surface charge on the cytotoxicity and uptake of carbon quantum dots in human umbilical cord derived mesenchymal stem cells. Colloids Surf. B Biointerfaces 2018, 171, 241–249. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, R.; Li, Q.; Maimaiti, T.; Lan, S.; Ouyang, P.; Ouyang, B.; Bai, Y.; Yu, B.; Yang, S.-T. Low toxicity of fluorescent carbon quantum dots to white rot fungus Phanerochaete chrysosporium. J. Environ. Chem. Eng. 2021, 9, 104633. [Google Scholar] [CrossRef]
- Xiao, Y.-Y.; Liu, L.; Chen, Y.; Zeng, Y.-L.; Liu, M.-Z.; Jin, L. Developmental toxicity of carbon quantum dots to the embryos/larvae of rare minnow (Gobiocypris rarus). BioMed Res. Int. 2016, 2016, 4016402. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Neuss, S.; Leifert, A.; Fischler, M.; Wen, F.; Simon, U.; Schmid, G.; Brandau, W.; Jahnen-Dechent, W. Size-dependent cytotoxicity of gold nanoparticles. Small 2007, 3, 1941–1949. [Google Scholar] [CrossRef]
- Navarro-Ruiz, M.C.; Cayuela, A.; Soriano, M.L.; Guzmán-Ruiz, R.; Malagón, M.M.; Valcárcel, M. A systematic comparative study of the toxicity of semiconductor and graphitic carbon-based quantum dots using in vitro cell models. Appl. Sci. 2020, 10, 8845. [Google Scholar] [CrossRef]
- Chung, C.Y.; Chen, Y.J.; Kang, C.H.; Lin, H.Y.; Huang, C.C.; Hsu, P.H.; Lin, H.J. Toxic or not toxic, that is the carbon quantum dot’s question: A comprehensive evaluation with zebrafish embryo, eleutheroembryo, and adult models. Polymers 2021, 13, 1598. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yao, J.; Jin, J.; Ma, J.; Masakorala, K. Microbial toxicity of a type of carbon dots to Escherichia coli. Arch. Environ. Contam. Toxicol. 2015, 69, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, S.; Cao, W.; Zhang, G.; Yang, X.; Gong, X.; Xing, X. Low-toxicity carbon quantum dots derived from gentamicin sulfate to combat antibiotic resistance and eradicate mature biofilms. Chem. Commun. 2020, 56, 2316–2319. [Google Scholar] [CrossRef]
- Arcudi, F.; Đorđević, L.; Prato, M. Synthesis, separation, and characterization of small and highly fluorescent nitrogen-doped carbon nanodots. Angew. Chem. Int. Ed. 2016, 55, 2107–2112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, A.A.; Mansour, M.K.; El Ahl, R.M.S.; El Hamaky, A.M.; Oraby, N.H. Toxic and beneficial effects of carbon nanomaterials on human and animal health. In Carbon Nanomaterials for Agri-Food and Environmental Applications; Elsevier: Amsterdam, The Netherlands, 2020; pp. 535–555. [Google Scholar]
- Jia, G.; Wang, H.; Yan, L.; Wang, X.; Pei, R.; Yan, T.; Zhao, Y.; Guo, X. Cytotoxicity of carbon nanomaterials: Single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol. 2005, 39, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Cheon, J.Y.; Kim, S.J.; Rhee, Y.H.; Kwon, O.H.; Park, W.H. Shape-dependent antimicrobial activities of silver nanoparticles. Int. J. Nanomed. 2019, 14, 2773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, F.; Lu, J.E.; Mercado, R.; Rojas-Andrade, M.D.; Ning, S.; Azhar, Z.; Sandhu, J.; Cazares, R.; Saltikov, C.; Chen, S. Antibacterial activity of nitrogen-doped carbon dots enhanced by atomic dispersion of copper. Langmuir 2020, 36, 11629–11636. [Google Scholar] [CrossRef]
- Wu, F.; Su, H.; Wang, K.; Wong, W.-K.; Zhu, X. Facile synthesis of N-rich carbon quantum dots from porphyrins as efficient probes for bioimaging and biosensing in living cells. Int. J. Nanomed. 2017, 12, 7375. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, M.; Bhandari, B.; Bai, B. Microbial and quality improvement of boiled gansi dish using carbon dots combined with radio frequency treatment. Int. J. Food Microbiol. 2020, 334, 108835. [Google Scholar] [CrossRef]
- Saravanan, A.; Maruthapandi, M.; Das, P.; Luong, J.H.; Gedanken, A. Green synthesis of multifunctional carbon dots with antibacterial activities. Nanomaterials 2021, 11, 369. [Google Scholar] [CrossRef]
- Gaweł, S.; Wardas, M.; Niedworok, E.; Wardas, P. Malondialdehyde (MDA) as a lipid peroxidation marker. Wiad. Lek. 2004, 57, 453–455. [Google Scholar]
- Wang, Z.; Zhu, X.; Su, Y.; Xu, W.; Liu, H.; Liu, Z.; Chen, W.; Wang, J. Dimethyl phthalate damaged the cell membrane of Escherichia coli K12. Ecotoxicol. Environ. Saf. 2019, 180, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Shang, E.; Niu, J.; Li, Y.; Zhou, Y.; Crittenden, J.C. Comparative toxicity of Cd, Mo, and W sulphide nanomaterials toward E. coli under UV irradiation. Environ. Pollut. 2017, 224, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.K.; Nenavathu, B.P.; Gangishetty, M.K.; Reddy, A. Studies on antibacterial activity of ZnO nanoparticles by ROS induced lipid peroxidation. Colloids Surf. B Biointerfaces 2012, 94, 143–150. [Google Scholar] [CrossRef]
- Oh, S.J.; Kim, H.; Liu, Y.; Han, H.K.; Kwon, K.; Chang, K.H.; Park, K.; Kim, Y.; Shim, K.; An, S.S.A. Incompatibility of silver nanoparticles with lactate dehydrogenase leakage assay for cellular viability test is attributed to protein binding and reactive oxygen species generation. Toxicol. Lett. 2014, 225, 422–432. [Google Scholar] [CrossRef]
- Devi, K.P.; Nisha, S.A.; Sakthivel, R.; Pandian, S.K. Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J. Ethnopharmacol. 2010, 130, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Bharali, P.; Konwar, B. Mode of antibacterial activity of eclalbasaponin isolated from Eclipta alba. Appl. Biochem. Biotechnol. 2013, 171, 2003–2019. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Kumar, S.; Tripathi, A.; Das, M.; Dwivedi, P.D. Interactive threats of nanoparticles to the biological system. Immunol. Lett. 2014, 158, 79–87. [Google Scholar] [CrossRef]
- Lin, N.; Berton, P.; Moraes, C.; Rogers, R.D.; Tufenkji, N. Nanodarts, nanoblades, and nanospikes: Mechano-bactericida nanostructures and where to find them. Adv. Colloid Interface Sci. 2018, 252, 55–68. [Google Scholar] [CrossRef] [Green Version]
- Qiang, S.; Li, Z.; Zhang, L.; Luo, D.; Geng, R.; Zeng, X.; Liang, J.; Li, P.; Fan, Q. Cytotoxic Effect of Graphene Oxide Nanoribbons on Escherichia coli. Nanomaterials 2021, 11, 1339. [Google Scholar] [CrossRef]
- Li, W.; Song, P.; Xin, Y.; Kuang, Z.; Liu, Q.; Ge, F.; Zhu, L.; Zhang, X.; Tao, Y. The effects of luminescent CdSe quantum dot-functionalized antimicrobial peptides nanoparticles on antibacterial activity and molecular mechanism. Int. J. Nanomed. 2021, 16, 1849. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Shen, J.; Liu, H.; Li, J.; Wang, A.; Hui, A.; Munir, H.A. Superoxide anion: Critical source of high performance antibacterial activity in Co-Doped ZnO QDs. Ceram. Int. 2020, 46, 15822–15830. [Google Scholar] [CrossRef]
- Ghosh, M.; Chakraborty, A.; Bandyopadhyay, M.; Mukherjee, A. Multi-walled carbon nanotubes (MWCNT): Induction of DNA damage in plant and mammalian cells. J. Hazard. Mater. 2011, 197, 327–336. [Google Scholar] [CrossRef]
- Domingues, M.M.; Silva, P.M.; Franquelim, H.G.; Carvalho, F.A.; Castanho, M.A.; Santos, N.C. Antimicrobial protein rBPI21-induced surface changes on Gram-negative and Gram-positive bacteria. Nanomediciine 2014, 10, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Maillard, A.P.F.; Espeche, J.C.; Maturana, P.; Cutro, A.C.; Hollmann, A. Zeta potential beyond materials science: Applications to bacterial systems and to the development of novel antimicrobials. Biochim. Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183597. [Google Scholar] [CrossRef]
- Halder, S.; Yadav, K.K.; Sarkar, R.; Mukherjee, S.; Saha, P.; Haldar, S.; Karmakar, S.; Sen, T. Alteration of Zeta potential and membrane permeability in bacteria: A study with cationic agents. SpringerPlus 2015, 4, 672. [Google Scholar] [CrossRef] [Green Version]
- Arakha, M.; Saleem, M.; Mallick, B.C.; Jha, S. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci. Rep. 2015, 5, 9578. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Jiang, F.; Xiao, Q.; Li, J.; Liu, X.; Yu, Q.; Liu, Y.; Zeng, C. Microcalorimetric, spectroscopic and microscopic investigation on the toxic effects of CdTe quantum dots on Halobacterium halobium R1. Nanotechnology 2010, 21, 475102. [Google Scholar] [CrossRef]
- Suescún-Bolívar, L.P.; Thomé, P.E. Osmosensing and osmoregulation in unicellular eukaryotes. World J. Microbiol. Biotechnol. 2015, 31, 435–443. [Google Scholar] [CrossRef]
- Yang, P.; Zhu, Z.; Zhang, T.; Zhang, W.; Chen, W.; Cao, Y.; Chen, M.; Zhou, X. Orange-emissive carbon quantum dots: Toward application in wound pH monitoring based on colorimetric and fluorescent changing. Small 2019, 15, 1902823. [Google Scholar] [CrossRef]
- Thoo, L.; Fahmi, M.Z.; Zulkipli, I.N.; Keasberry, N.; Idris, A. Interaction and cellular uptake of surface-modified carbon dot nanoparticles by J774. 1 macrophages. Cent. Eur. J. Immunol. 2017, 42, 324–330. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiang, S.; Zhang, L.; Li, Z.; Liang, J.; Li, P.; Song, J.; Guo, K.; Wang, Z.; Fan, Q. New Insights into the Cellular Toxicity of Carbon Quantum Dots to Escherichia coli. Antioxidants 2022, 11, 2475. https://doi.org/10.3390/antiox11122475
Qiang S, Zhang L, Li Z, Liang J, Li P, Song J, Guo K, Wang Z, Fan Q. New Insights into the Cellular Toxicity of Carbon Quantum Dots to Escherichia coli. Antioxidants. 2022; 11(12):2475. https://doi.org/10.3390/antiox11122475
Chicago/Turabian StyleQiang, Shirong, Li Zhang, Zhengbin Li, Jianjun Liang, Ping Li, Jiayu Song, Kunling Guo, Zihuan Wang, and Qiaohui Fan. 2022. "New Insights into the Cellular Toxicity of Carbon Quantum Dots to Escherichia coli" Antioxidants 11, no. 12: 2475. https://doi.org/10.3390/antiox11122475



