Impact of Hydrogen Peroxide on Protein Synthesis in Yeast
Abstract
:1. Introduction
2. Systems Involved in Responses to H2O2
3. Redox Regulation of Protein Synthesis
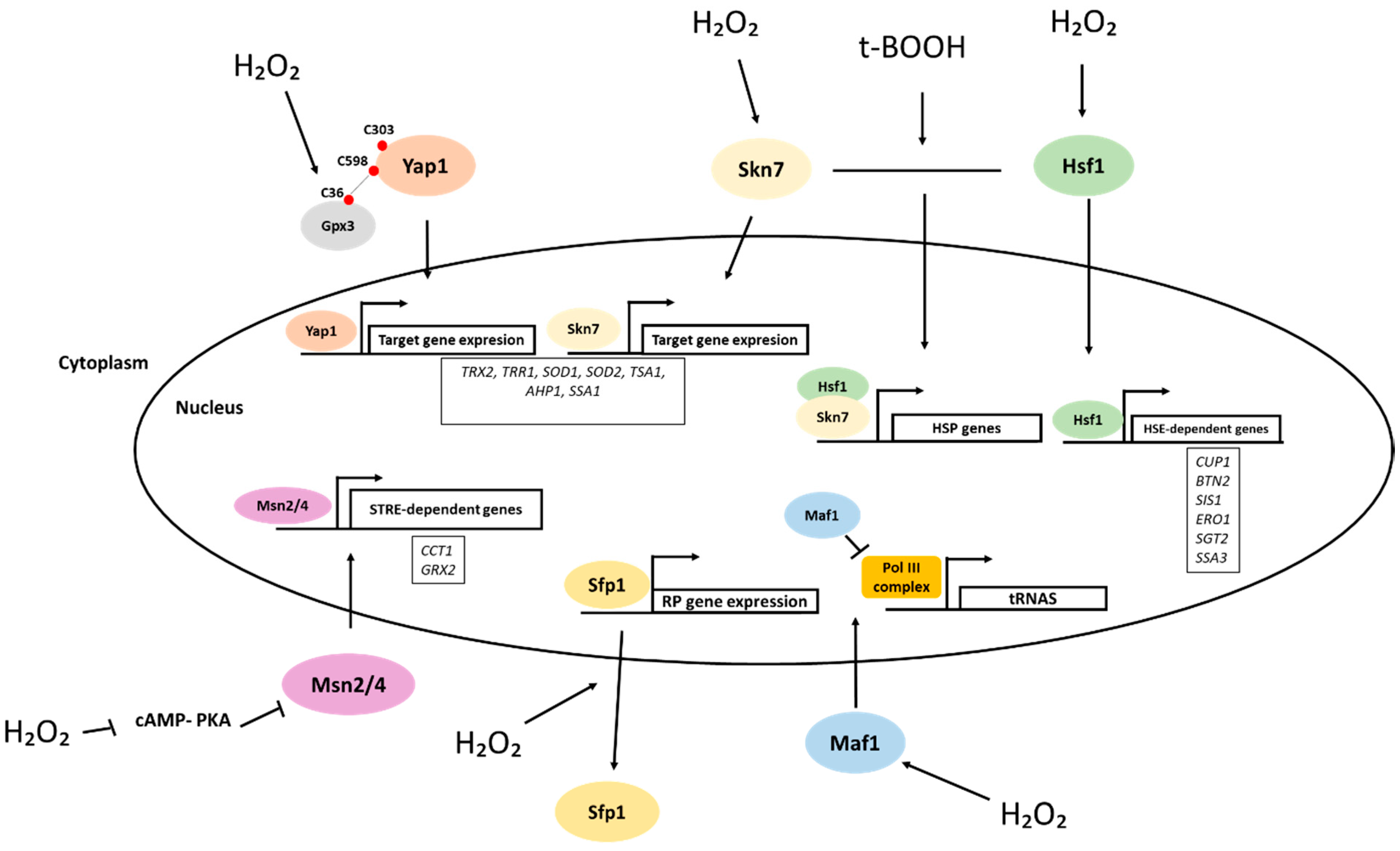
4. Role of H2O2 in Protein Synthesis: Transcription
5. Roles of H2O2 in Protein Synthesis: Translation
5.1. Translation Initiation
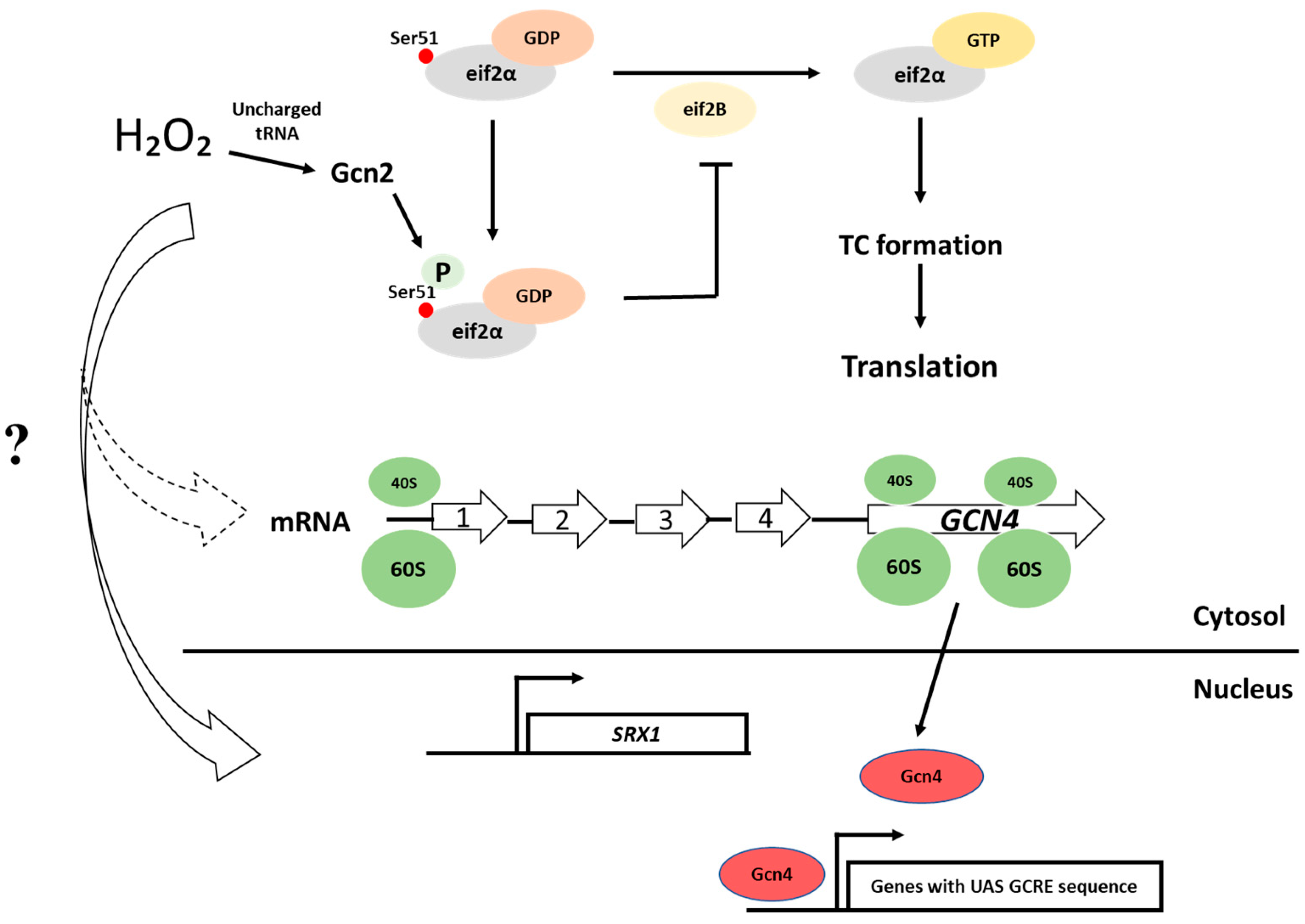
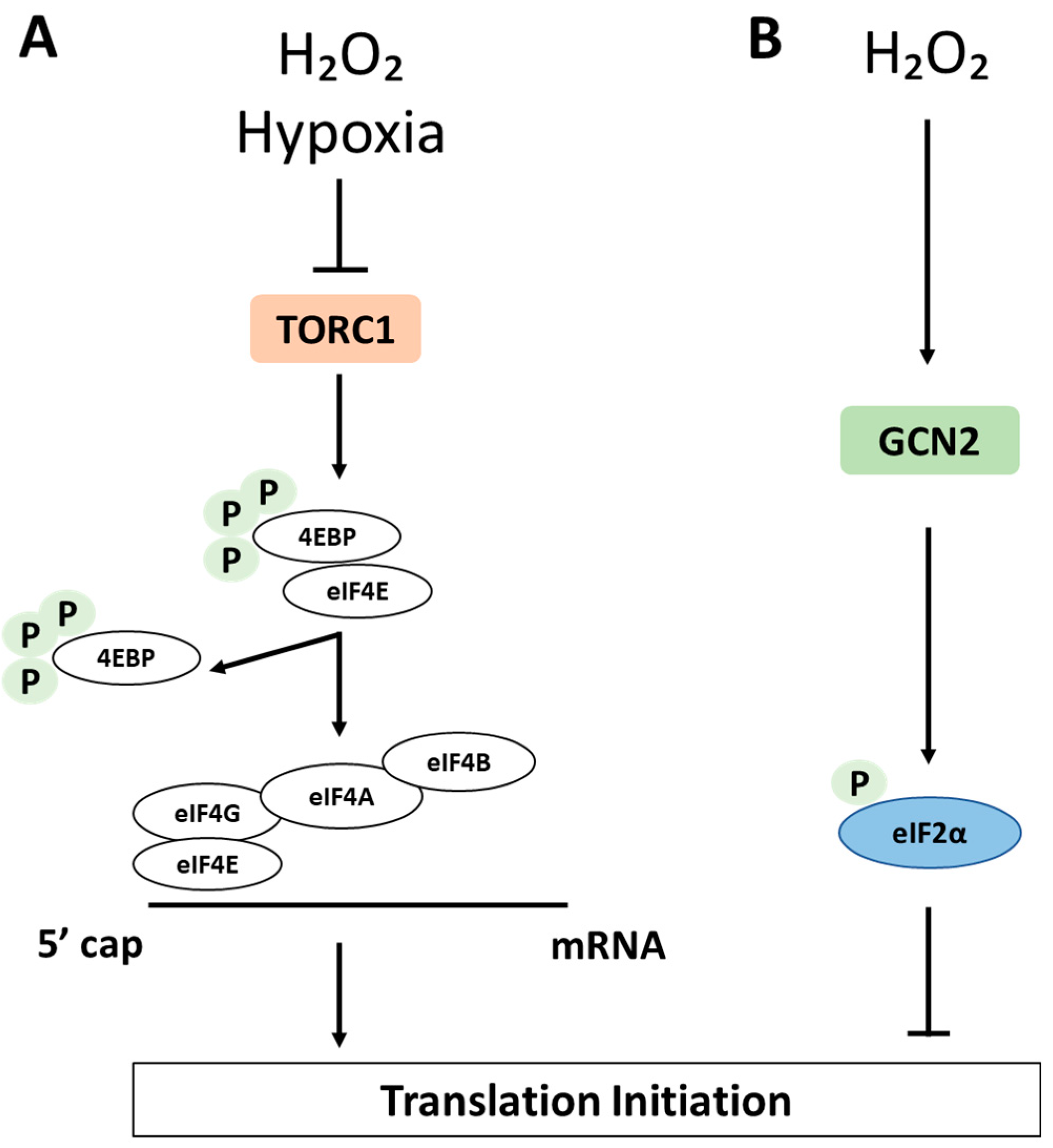
5.1.1. The Integrated Stress Response
5.1.2. The TOR Pathway
5.1.3. Mitochondrial Stress Response Pathways
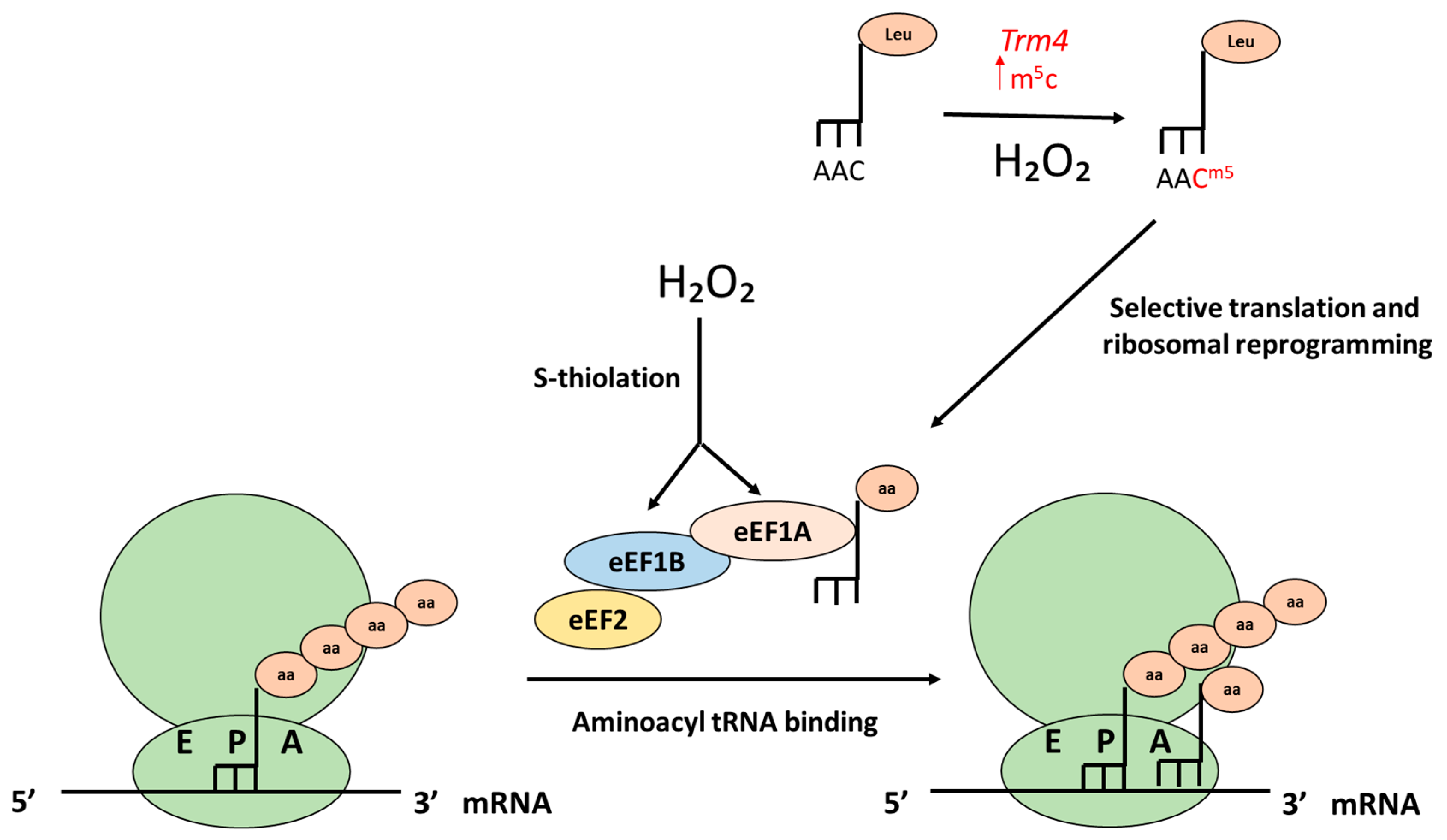
5.2. Translation Elongation
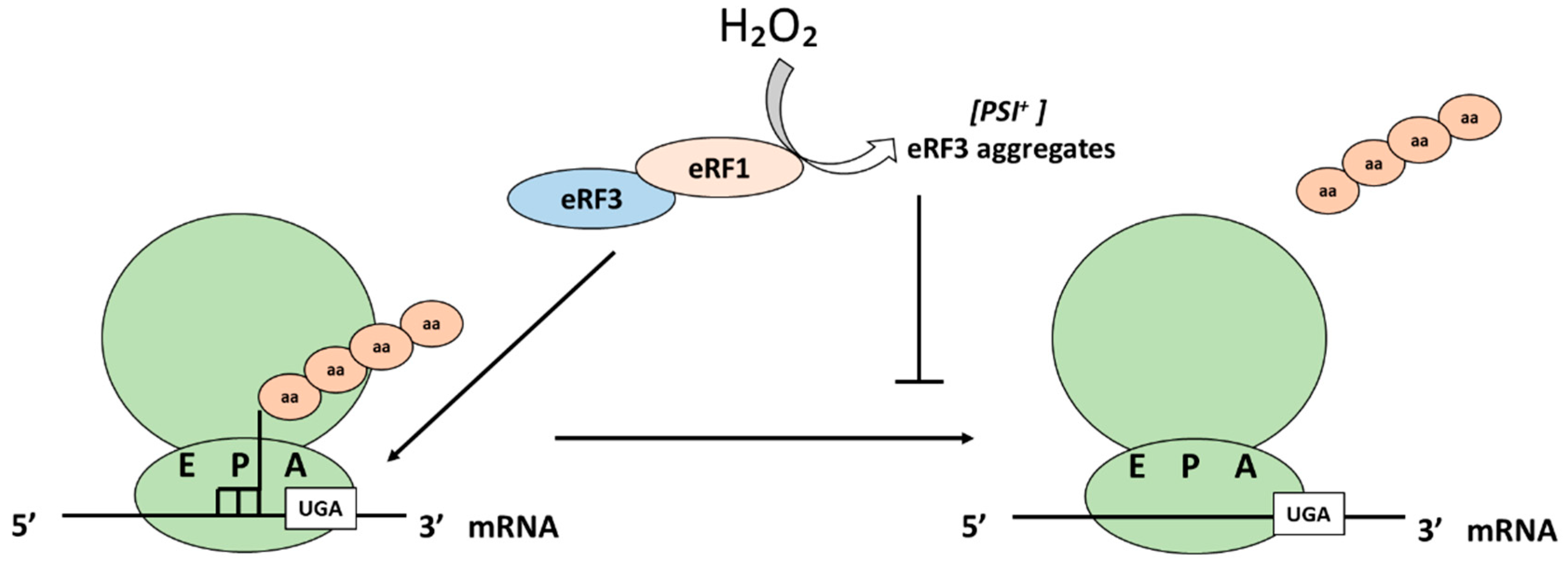
5.3. Translation Termination
5.4. Ribosomal Recycling
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Davies, K.J. Oxidative stress: The paradox of aerobic life. Biochem. Soc. Symp. 1995, 61, 1–31. [Google Scholar] [PubMed]
- Murphy, M.P.; Holmgren, A.; Larsson, N.G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Lushchak, V.I. Chemico-Biological Interactions Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014, 224, 164–175. [Google Scholar] [CrossRef]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [Green Version]
- Go, Y.; Jones, D.P. The Redox Proteome. J. Biol. Chem. 2013, 288, 26512–26520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korovila, I.; Hugo, M.; Castro, J.P.; Weber, D.; Höhn, A.; Grune, T.; Jung, T. Proteostasis, oxidative stress and aging. Redox Biol. 2017, 13, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Ristow, M.; Schmeisser, S. Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 2011, 51, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Bodvard, K.; Peeters, K.; Roger, F.; Romanov, N.; Igbaria, A.; Welkenhuysen, N.; Palais, G.; Reiter, W.; Toledano, M.B.; Käll, M.; et al. Light-sensing via hydrogen peroxide and a peroxiredoxin. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Amponsah, P.S.; Yahya, G.; Zimmermann, J.; Mai, M.; Mergel, S.; Mühlhaus, T.; Storchova, Z.; Morgan, B. Peroxiredoxins couple metabolism and cell division in an ultradian cycle. Nat. Chem. Biol. 2021, 17, 477–484. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 1989, 264, 13963–13966. [Google Scholar] [CrossRef]
- Toledano, M.B.; Kumar, C.; Le Moan, N.; Spector, D.; Tacnet, F. The system biology of thiol redox system in Escherichia coli and yeast: Differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett. 2007, 581, 3598–3607. [Google Scholar] [CrossRef] [Green Version]
- Penninckx, M.J. An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2002, 2, 295–305. [Google Scholar]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Le Moan, N.; Clement, G.; Le Maout, S. The Saccharomyces cerevisiae Proteome of Oxidized Protein Thiols: Contrasted functions for the thioredoxin and glutathione pathways. J. Biol. Chem. 2006, 281, 10420–10430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, C.; Igbaria, A.; D’Autreaux, B.; Planson, A.G.; Junot, C.; Godat, E.; Bachhawat, A.K.; Delaunay-Moisan, A.; Toledano, M.B. Glutathione revisited: A vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 2011, 30, 2044–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaunay, A.; Pflieger, D.; Barrault, M.B.; Vinh, J.; Toledano, M.B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 2002, 111, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Nyström, T.; Yang, J.; Molin, M. Peroxiredoxins, gerontogenes linking aging to genome instability and cancer. Genes Dev. 2012, 26, 2001–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molin, M.; Troussicot, L.; Troussicot, L.; Burmann, B.M.; Molin, M. Structure Structural determinants of multimerization and dissociation in 2-Cys peroxiredoxin chaperone function. Structure 2021. [Google Scholar] [CrossRef]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Rhee, S.G.; Kil, I.S. Multiple Functions and Regulation of Mammalian Peroxiredoxins. Annu. Rev. Biochem. 2017, 86, 749–775. [Google Scholar] [CrossRef]
- Wood, Z.; Poole, L.; Karplus, A. Peroxiredoxin Evolution and the Signaling. Science 2003, 300, 650–654. [Google Scholar] [CrossRef]
- Woo, H.A.; Yim, S.H.; Shin, D.H.; Kang, D.; Yu, D.Y.; Rhee, S.G. Inactivation of Peroxiredoxin I by Phosphorylation Allows Localized H2O2 Accumulation for Cell Signaling. Cell 2010, 140, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Goulev, Y.; Morlot, S.; Matifas, A.; Huang, B.; Molin, M.; Toledano, M.B.; Charvin, G. Nonlinear feedback drives homeostatic plasticity in H2O2 stress response. Elife 2017, 6, e23971. [Google Scholar] [CrossRef]
- Roger, F.; Picazo, C.; Asami, C.; Reiter, W.; Hanzén, S.; Gao, C.; Lagniel, G.; Welkenhuysen, N.; Labarre, J.; Nyström, T.; et al. Peroxiredoxin promotes longevity and H2O2-resistance in yeast through redox-modulation of protein kinase a. Elife 2020, 9, e60346. [Google Scholar] [CrossRef] [PubMed]
- Stöcker, S.; Maurer, M.; Ruppert, T.; Dick, T.P. A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol. 2018, 14, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, T.J.; Lee, K. A novel function of peroxiredoxin 1 (Prx-1) in apoptosis signal-regulating kinase 1 (ASK1)-mediated signaling pathway. FEBS Lett. 2008, 582, 1913–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Sabharwal, P.; Rao, M.; Sockanathan, S. The Antioxidant Enzyme Prdx1 Controls Neuronal Differentiation by Thiol-Redox-Dependent Activation of GDE2. Cell 2009, 138, 1209–1221. [Google Scholar] [CrossRef] [Green Version]
- Hanzén, S.; Vielfort, K.; Yang, J.; Roger, F.; Andersson, V.; Zamarbide-Forés, S.; Andersson, R.; Malm, L.; Palais, G.; Biteau, B.; et al. Lifespan Control by Redox-Dependent Recruitment of Chaperones to Misfolded Proteins. Cell 2016, 166, 140–151. [Google Scholar] [CrossRef] [Green Version]
- Proud, C.G. eIF2 and the control of cell physiology. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2005; Volume 16, pp. 3–12. [Google Scholar]
- Clemens, M.J. Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J. Cell. Mol. Med. 2001, 5, 221–239. [Google Scholar] [CrossRef]
- Crespo, J.L.; Powers, T.; Fowler, B.; Hall, M.N. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 2002, 99, 6784–6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushchak, O.; Strilbytska, O.; Piskovatska, V.; Storey, K.B.; Koliada, A.; Vaiserman, A. The role of the tor pathway in mediating the link between nutrition and longevity. Mech. Ageing Dev. 2017, 164, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032607. [Google Scholar] [CrossRef]
- Advani, V.M.; Ivanov, P. Translational Control under Stress: Reshaping the Translatome. BioEssays 2019, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Hussmann, J.A.; Weissman, J.S. Ribosome profiling: Global views of translation. Cold Spring Harb. Perspect. Biol. 2019, 11, 1–20. [Google Scholar] [CrossRef]
- Gingold, H.; Tehler, D.; Christoffersen, N.R.; Nielsen, M.M.; Asmar, F.; Kooistra, S.M.; Christophersen, N.S.; Christensen, L.L.; Borre, M.; Sørensen, K.D.; et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014, 158, 1281–1292. [Google Scholar] [CrossRef] [Green Version]
- Topf, U.; Suppanz, I.; Samluk, L.; Wrobel, L.; Böser, A.; Sakowska, P.; Knapp, B.; Pietrzyk, M.K.; Chacinska, A.; Warscheid, B. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Grant, C.M. Regulation of translation by hydrogen peroxide. Antioxid. Redox Signal. 2011, 15, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, Á.; Isnard, A. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000, 19, 5157–5166. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 Control Two Specialized Oxidative Stress Response Regulons in Yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef] [Green Version]
- Beckhouse, A.G.; Grant, C.M.; Rogers, P.J.; Dawes, I.W.; Higgins, V.J. The adaptive response of anaerobically grown Saccharomyces cerevisiae to hydrogen peroxide is mediated by the Yap1 and Skn7 transcription factors. FEMS Yeast Res. 2008, 8, 1214–1222. [Google Scholar] [CrossRef] [Green Version]
- Barna, J.; Csermely, P.; Vellai, T. Roles of heat shock factor 1 beyond the heat shock response. Cell. Mol. Life Sci. 2018, 75, 2897–2916. [Google Scholar] [CrossRef]
- Davidson, J.F.; Whyte, B.; Bissinger, P.H.; Schiestl, R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, K.; Kawamura, A.; Izawa, S.; Inoue, Y. Role of glutathione in heat-shock-induced cell death of Saccharomyces cerevisiae. Biochem. J. 2000, 352, 71–78. [Google Scholar] [CrossRef]
- Yamamoto, A.; Ueda, J.; Yamamoto, N.; Hashikawa, N.; Sakurai, H. Role of Heat Shock Transcription Factor in Saccharomyces cerevisiae Oxidative Stress Response. Eukaryot. Cell 2007, 6, 1373–1379. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.; Thiele, D.J. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003, 17, 516–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masser, A.E.; Kang, W.; Roy, J.; Kaimal, J.M.; Quintana-Cordero, J.; Friedländer, M.R.; Andréasson, C. Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1. Elife 2019, 8, e47791. [Google Scholar] [CrossRef]
- Wang, Y.; Gibney, P.A.; West, J.D.; Morano, K.A. The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds. Mol. Biol. Cell 2012, 23, 3290–3298. [Google Scholar] [CrossRef]
- Raitt, D.C.; Johnson, A.L.; Erkine, A.M.; Makino, K.; Morgan, B.; Gross, D.S.; Johnston, L.H. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 2000, 11, 2335–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagniel, G.; Garmendia-torres, C.; Molin, M.; Boy-marcotte, E.; Jacquet, M.; Toledano, M.B.; Labarre, J. H2O2 Activates the Nuclear Localization of Msn2 and Maf1 through Thioredoxins in Saccharomyces cerevisiae. Eurayot. Cell 2009, 8, 1429–1438. [Google Scholar]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sobotta, M.C.; Liou, W.; Stöcker, S.; Talwar, D.; Oehler, M.; Ruppert, T.; Scharf, A.N.D.; Dick, T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015, 11, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Marion, R.M.; Regev, A.; Segal, E.; Barash, Y.; Koller, D.; Friedman, N.; Shea, E.K.O. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 2004, 101, 14315–14322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavitt, G.D.; Ramaiah, K.V.A.; Kimball, S.R.; Hinnebusch, A.G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine–nucleotide exchange. Genes Dev. 1998, 12, 514–526. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, K.; Meyer, M.R.; Belinda, M.; Slade, D.; Roberts, C.; Alan, G.; Marton, M.J.; Jackson, B.M.; Hinnebusch, A.G. Transcriptional Profiling Shows that Gcn4p Is a Master Regulator of Gene Expression during Amino Acid Starvation in Yeast. Mol. Cell. Biol. 2001, 21, 4347–4368. [Google Scholar] [CrossRef] [Green Version]
- Dever, T.E. Gene-specific regulation by general translation factors. Cell 2002, 108, 545–556. [Google Scholar] [CrossRef] [Green Version]
- Shenton, D.; Smirnova, J.B.; Selley, J.N.; Carroll, K.; Hubbard, S.J.; Pavitt, G.D.; Ashe, M.P.; Grant, C.M. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006, 281, 29011–29021. [Google Scholar] [CrossRef] [Green Version]
- Shenton, D.; Grant, C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003, 374, 513–519. [Google Scholar] [CrossRef] [Green Version]
- Molin, M.; Yang, J.; Hanzen, S.; Toledano, M.B.; Labarre, J.; Nystrom, T. Life span extension and H2O2 resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol. Cell 2011, 43, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Gast, V.; Campbell, K.; Picazo, C.; Engqvist, M.; Siewers, V.; Molin, M. The yeast eIF2 kinase Gcn2 facilitates H2O2-mediated feedback inhibition of both protein synthesis and ER oxidative folding during recombinant protein production. Appl. Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, M.V.; Lobanov, A.V.; Gladyshev, V.N. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 2012, 109, 17394–17399. [Google Scholar] [CrossRef] [Green Version]
- Baek, J.Y.; Han, S.H.; Sung, S.H.; Lee, H.E.; Kim, Y.M.; Noh, Y.H.; Bae, S.H.; Rhee, S.G.; Chang, T.S. Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J. Biol. Chem. 2012, 287, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebauer, F.; Hentze, M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004, 5, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Averous, J.; Proud, C.G. When translation meets transformation: The mTOR story. Oncogene 2006, 25, 6423–6435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonskikh, Y.; Polacek, N. Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 2017, 168, 30–36. [Google Scholar] [CrossRef]
- Liu, L.; Wise, D.R.; Diehl, J.A.; Simon, M.C. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J. Biol. Chem. 2008, 283, 31153–31162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topf, U.; Uszczynska-Ratajczak, B.; Chacinska, A. Mitochondrial stress-dependent regulation of cellular protein synthesis. J. Cell Sci. 2019, 132, jcs226258. [Google Scholar] [CrossRef] [Green Version]
- Wrobel, L.; Topf, U.; Bragoszewski, P.; Wiese, S.; Sztolsztener, M.E.; Oeljeklaus, S.; Varabyova, A.; Lirski, M.; Chroscicki, P.; Mroczek, S.; et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015, 524, 485–488. [Google Scholar] [CrossRef]
- Samluk, L.; Urbanska, M.; Kisielewska, K.; Mohanraj, K.; Kim, M.J.; Machnicka, K.; Liszewska, E.; Jaworski, J.; Chacinska, A. Cytosolic translational responses differ under conditions of severe short-term and long-term mitochondrial stress. Mol. Biol. Cell 2019, 30, 1864–1877. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. The mTOR pathway in the control of protein synthesis. Physiology 2006, 21, 362–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Showkat, M.; Beigh, M.A.; Andrabi, K.I. mTOR Signaling in Protein Translation Regulation: Implications in Cancer Genesis and Therapeutic Interventions. Mol. Biol. Int. 2014, 2014, 686984. [Google Scholar] [CrossRef] [Green Version]
- Rubio, A.; Ghosh, S.; Mülleder, M.; Ralser, M.; Mata, J. Ribosome profiling reveals ribosome stalling on tryptophan codons and ribosome queuing upon oxidative stress in fission yeast. Nucleic Acids Res. 2021, 49, 383–399. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; De Crécy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.O.; Esberg, A.; Huang, B.; Björk, G.R.; Byström, A.S. Eukaryotic Wobble Uridine Modifications Promote a Functionally Redundant Decoding System. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Laxman, S. tRNA wobble-uridine modifications as amino acid sensors and regulators of cellular metabolic state. Curr. Genet. 2020, 66, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Sideri, T.C.; Stojanovski, K.; Tuite, M.F.; Grant, C.M. Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 6394–6399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buskirk, A.R.; Green, R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youngman, E.M.; McDonald, M.E.; Green, R. Peptide release on the ribosome: Mechanism and implications for translational control. Annu. Rev. Microbiol. 2008, 62, 353–373. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.J.; Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, E1392–E1398. [Google Scholar] [CrossRef] [Green Version]
- Alhebshi, A.; Sideri, T.C.; Holland, S.L.; Avery, S.V. The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Mol. Biol. Cell 2012, 23, 3582–3590. [Google Scholar] [CrossRef] [PubMed]
- Young, D.J.; Guydosh, N.R.; Zhang, F.; Hinnebusch, A.G.; Green, R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3′UTRs In Vivo. Cell 2015, 162, 872–884. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.C.; Bootman, M.D.; Scott, J. Second messengers. Cold Spring Harb. Perspect. Biol. 2016, 8, 1–14. [Google Scholar] [CrossRef]
- Gorelick, F.S. Second messenger systems and adaptation. Gut 1987, 28, 79–84. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picazo, C.; Molin, M. Impact of Hydrogen Peroxide on Protein Synthesis in Yeast. Antioxidants 2021, 10, 952. https://doi.org/10.3390/antiox10060952
Picazo C, Molin M. Impact of Hydrogen Peroxide on Protein Synthesis in Yeast. Antioxidants. 2021; 10(6):952. https://doi.org/10.3390/antiox10060952
Chicago/Turabian StylePicazo, Cecilia, and Mikael Molin. 2021. "Impact of Hydrogen Peroxide on Protein Synthesis in Yeast" Antioxidants 10, no. 6: 952. https://doi.org/10.3390/antiox10060952




