Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC
Abstract
:1. Introduction
2. Materials and Methods
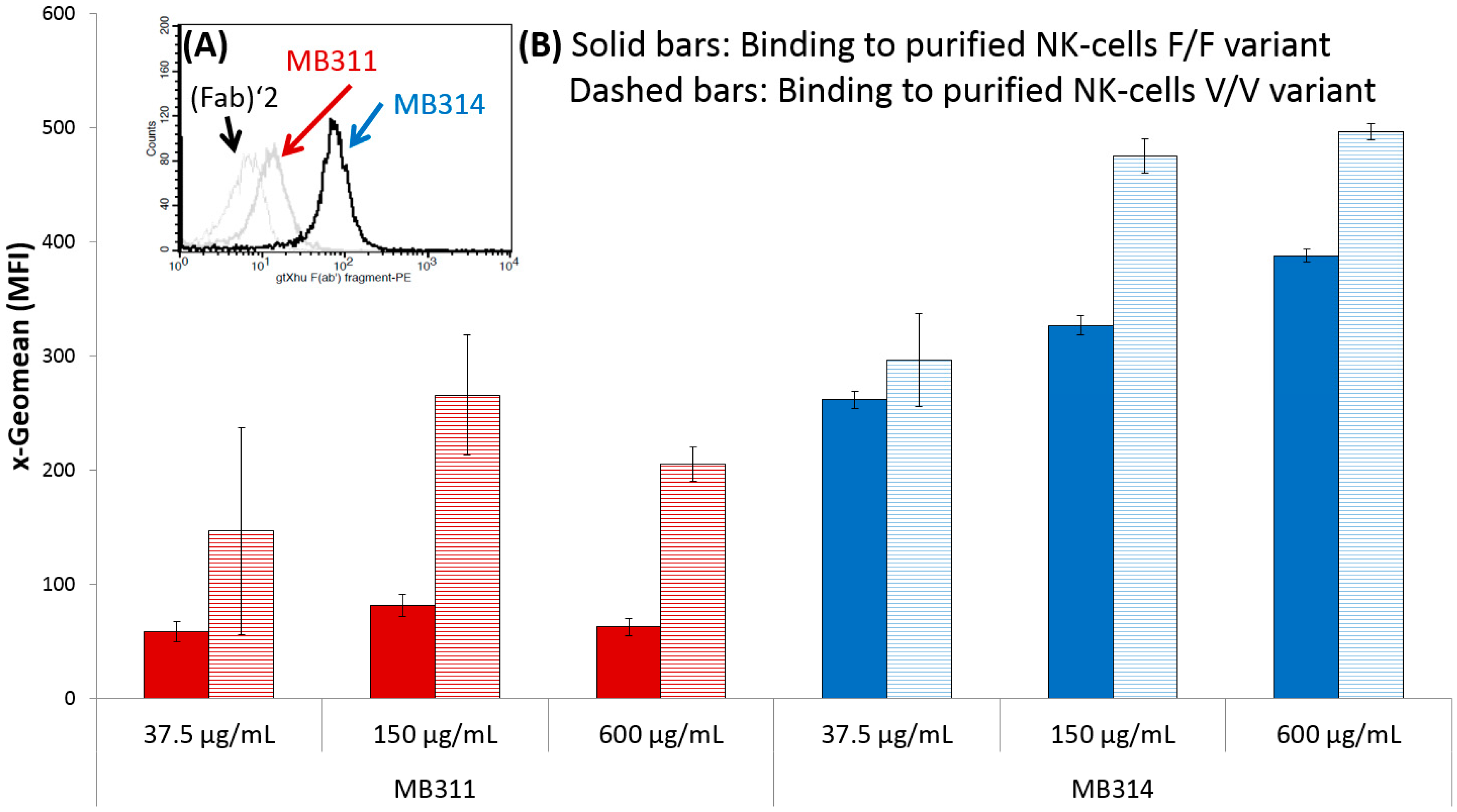
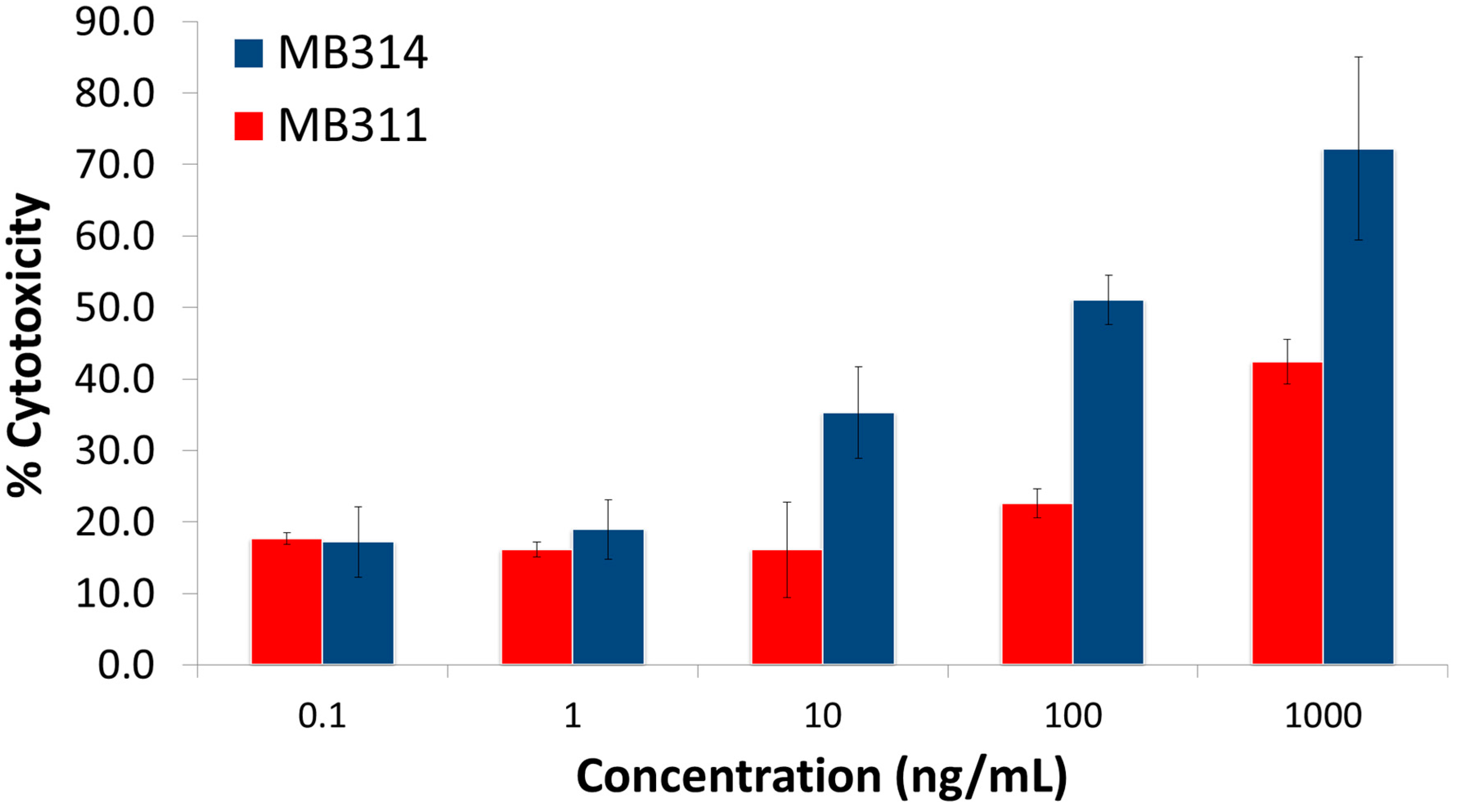
3. Results and Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Essentials in Gycobiology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2009. [Google Scholar]
- Huang, G.; Chen, X.; Xiao, F. New fabrication and applications of carbohydrate arrays. Curr. Med. Chem. 2014, 21, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, S.; Zhang, B. Glycan analysis of therapeutic glycoproteins. mAbs 2016, 8, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, S.; Zhang, B. The use of lectin microarray for assessing glycosylation of therapeutic proteins. mAbs 2016, 8, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Lux, A.; Nimmerjahn, F. Impact of differential glycosylation on IgG activity. Adv. Exp. Med. Biol. 2011, 780, 113–124. [Google Scholar] [PubMed]
- Yamane-Ohnuki, N.; Satoh, M. Production of therapeutic antibodies with controlled fucosylation. mAbs 2009, 1, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, C.; Grau, S.; Jäger, C.; Sondermann, P.; Brünker, P.; Waldhauer, I.; Hennig, M.; Ruf, A.; Rufer, A.C.; Stihle, M.; et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. USA 2011, 108, 12669–12674. [Google Scholar] [CrossRef] [PubMed]
- Listinsky, J.J.; Siegal, G.P.; Listinsky, C.M. Glyco-engineering in cancer therapeutics: A review with fucose-depleted trastuzumab as the model. Anticancer Drugs 2013, 24, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.; Liu, W.; Ng, T.B. Development and applications of lectins as biological tools in biomedical research. Med. Res. Rev. 2016, 36, 221–247. [Google Scholar] [CrossRef] [PubMed]
- Kuno, A.; Uchiyama, N.; Koseki-Kuno, S.; Ebe, Y.; Takashima, S.; Yamada, M.; Hirabayashi, J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat. Methods 2005, 2, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Kuno, A.; Tateno, H. Development and applications of the lectin microarray. Top. Curr. Chem. 2015, 367, 105–124. [Google Scholar] [PubMed]
- Oruzio, D.; Waxenecker, G.; Aulmann, C.; Märkl, B.; Wagner, T.; Mudde, G.; Schuster, M.; Eller, N.; Mayer, A.; Stranner, S.; et al. A. phase I dose escalation study with the Lewis Y carbohydrate specific humanized antibody IGN311. J. Cancer Ther. 2011, 2, 760–771. [Google Scholar] [CrossRef]
- Nechansky, A.; Schuster, M.; Jost, W.; Siegl, P.; Wiederkum, S.; Gorr, G.; Kircheis, R. Compensation of endogenous IgG mediated inhibition of antibody-dependent cellular cytotoxicity by glyco-engineering of therapeutic antibodies. Mol. Immunol. 2007, 44, 1815–1817. [Google Scholar] [CrossRef] [PubMed]
- Koene, H.R.; Kleijer, M.; Algra, J.; Roos, D.; von dem Borne, A.E.G.K.; de Haas, M. FcgRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell FcgRIIIa, independently of the FcgRIIIa-48L/R/H phenotype. Blood 1997, 90, 1109–1114. [Google Scholar] [PubMed]
- Kuno, A.K.; Itakura, Y.; Toyoda, M.; Takahashi, Y.; Yamada, M.; Umezawa, A.; Hirabayashi, J. Development of a data-mining system for differential profiling of cell glycoproteins based on lectin microarray. J. Proteom. Bioinform. 2008, 1, 068–072. [Google Scholar] [CrossRef]
- Tateno, H.; Nakamura-Tsuruta, S.; Hirabayashi, J. Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology 2009, 19, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Jost, W.; Mudde, G.C.; Wiederkum, S.; Schwager, C.; Janzek, E.; Altmann, F.; Stadlmann, J.; Stemmer, C.; Gorr, G. In vivo glyco-engineered antibody with improved lytic potential produced by an innovative non-mammalian expression system. Biotechnol. J. 2007, 2, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tao, S.C.; Bova, G.S.; Liu, A.Y.; Chan, D.W.; Zhu, H.; Zhang, H. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin based immunosorbent assays. Anal. Chem. 2011, 183, 8509–8516. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roucka, M.; Zimmermann, K.; Fido, M.; Nechansky, A. Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC. Microarrays 2017, 6, 1. https://doi.org/10.3390/microarrays6010001
Roucka M, Zimmermann K, Fido M, Nechansky A. Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC. Microarrays. 2017; 6(1):1. https://doi.org/10.3390/microarrays6010001
Chicago/Turabian StyleRoucka, Markus, Klaus Zimmermann, Markus Fido, and Andreas Nechansky. 2017. "Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC" Microarrays 6, no. 1: 1. https://doi.org/10.3390/microarrays6010001




