Tissue Microarray Technology for Molecular Applications: Investigation of Cross-Contamination between Tissue Samples Obtained from the Same Punching Device
Abstract
:1. Introduction
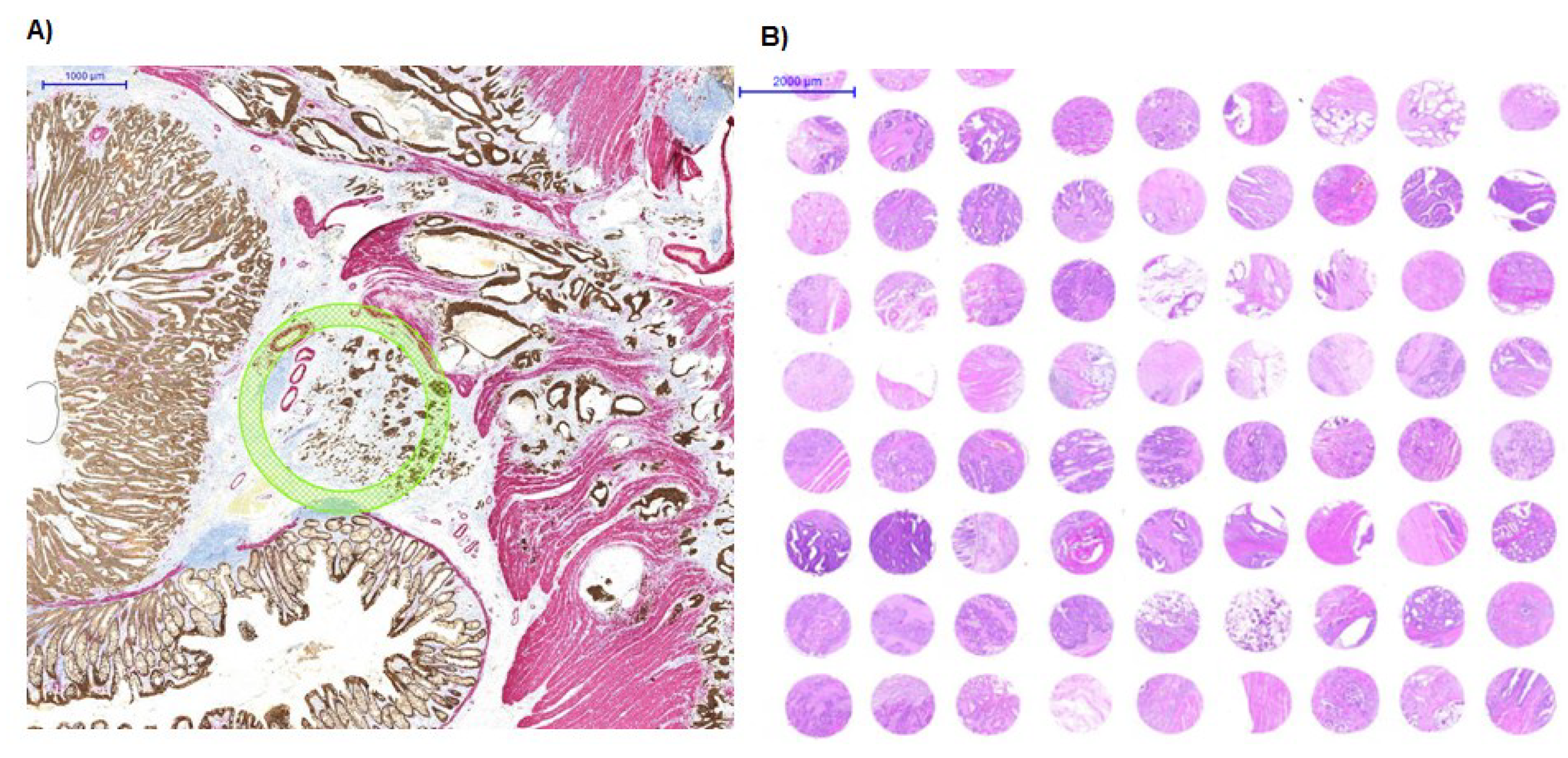
2. Experimental Design
2.1. Patients
2.2. Analysis of Alternating Mycobacterium Positive and Negative Tissue Samples
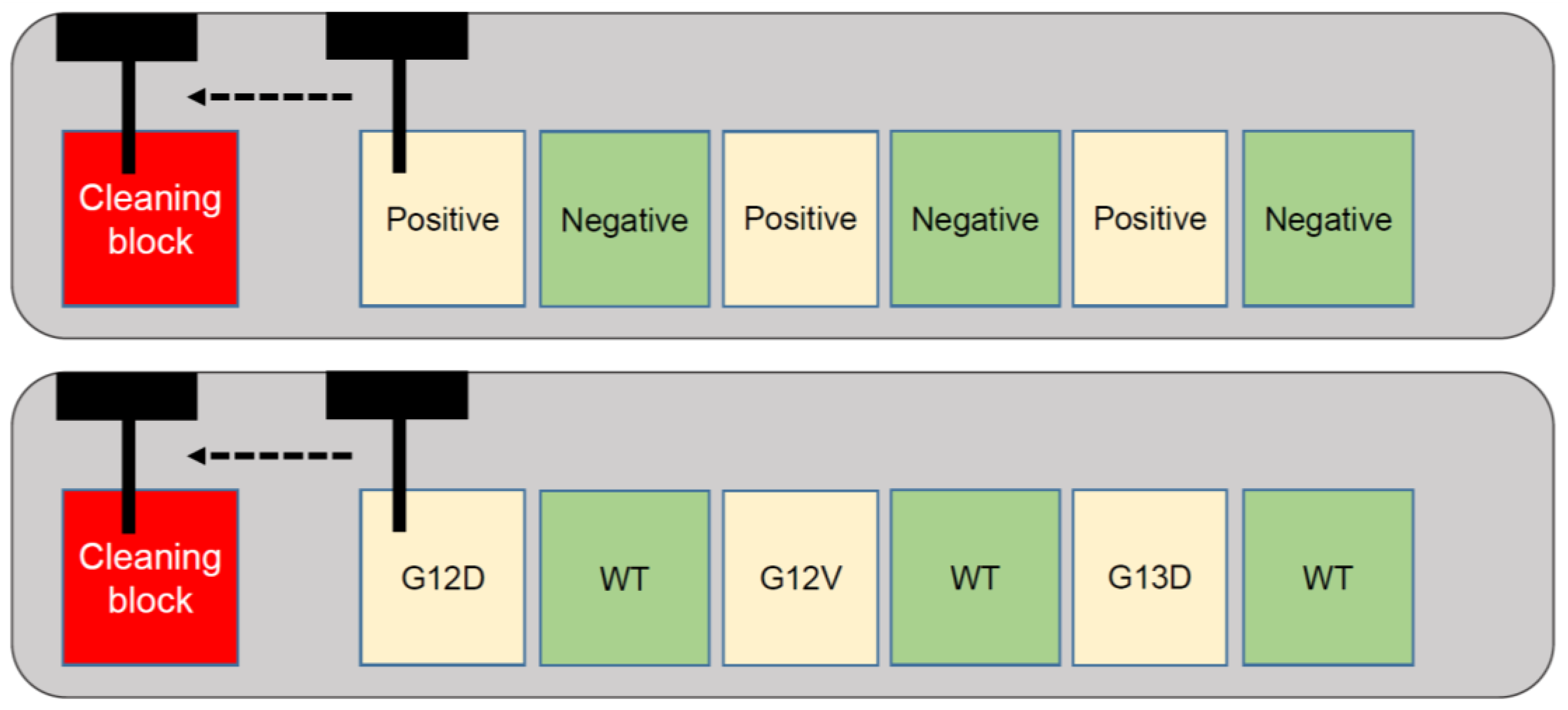
2.3. Analysis of Alternating KRAS Mutated and Wild-Type Tissue Samples
3. Results/Discussion
3.1. An Alternate Use of TMA Technology
3.2. Cross-Contamination Assessed Using Mycobacterium Tuberculosis Assay
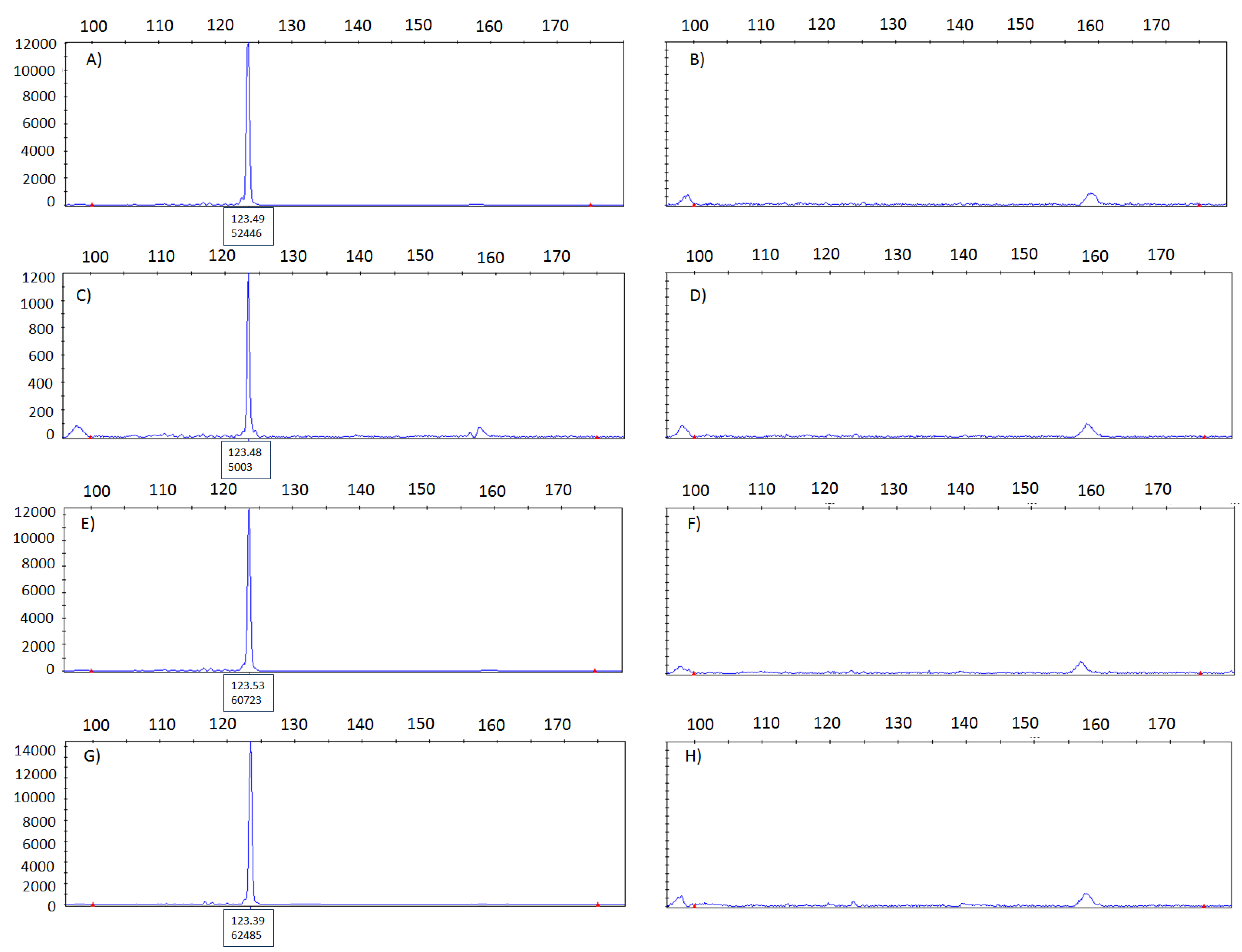
3.3. Cross-Contamination Assessed Using KRAS Analysis by Pyrosequencing
3.4. Impact of the Finding
3.5. Conclusion

Author Contributions
Conflicts of Interest
References
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Kononen, J.; Koivisto, P.; Schraml, P.; Moch, H.; Gasser, T.C.; Willi, N.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999, 59, 803–806. [Google Scholar]
- Hsu, F.D.; Nielsen, T.O.; Alkushi, A.; Dupuis, B.; Huntsman, D.; Liu, C.L.; van de Rijn, M.; Gilks, C.B. Tissue microarrays are an effective quality assurance tool for diagnostic immunohistochemistry. Mod. Pathol. 2002, 15, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Voduc, D.; Kenney, C.; Nielsen, T.O. Tissue microarrays in clinical oncology. Semin. Radiat. Oncol. 2008, 18, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Bordeaux, J.M.; Cheng, H.; Welsh, A.W.; Haffty, B.G.; Lannin, D.R.; Wu, X.; Su, N.; Ma, X.J.; Luo, Y.; Rimm, D.L. Quantitative in situ measurement of estrogen receptor mRNA predicts response to tamoxifen. PLoS ONE 2012, 7, e36559. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.D.; Jones, M.A.; Beadle, G.F.; Stein, S.R. Bright-field in situ hybridization for HER2 gene amplification in breast cancer using tissue microarrays: Correlation between chromogenic (CISH) and automated silver-enhanced (SISH) methods with patient outcome. Diagn. Mol. Pathol. 2009, 18, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Terracciano, L.; Tornillo, L.; Gunthert, U.; Vuong, T.; Jass, J.R.; Lugli, A. Role of RHAMM within the hierarchy of well-established prognostic factors in colorectal cancer. Gut 2008, 57, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Ballman, K.; Vassilakopoulou, M.; Dueck, A.C.; Reinholz, M.M.; Tenner, K.; Gralow, J.; Hudis, C.; Davidson, N.E.; Fountzilas, G.; et al. EGFR expression is associated with decreased benefit from trastuzumab in the NCCTG N9831 (Alliance) trial. Br. J. Cancer 2014, 111, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Suter, G.; Perren, A.; Lugli, A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. J. Vis. Exp. 2014, 23, e51893–e51893. [Google Scholar]
- Zlobec, I.; Koelzer, V.H.; Dawson, H.; Perren, A.; Lugli, A. Next-generation tissue microarray (ngTMA) increases the quality of biomarker studies: An example using CD3, CD8, and CD45RO in the tumor microenvironment of six different solid tumor types. J. Transl. Med. 2013, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Thierry, D.; Brisson-Noel, A.; Vincent-Levy-Frebault, V.; Nguyen, S.; Guesdon, J.L.; Gicquel, B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 1990, 28, 2668–2673. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vassella, E.; Galván, J.A.; Zlobec, I. Tissue Microarray Technology for Molecular Applications: Investigation of Cross-Contamination between Tissue Samples Obtained from the Same Punching Device. Microarrays 2015, 4, 188-195. https://doi.org/10.3390/microarrays4020188
Vassella E, Galván JA, Zlobec I. Tissue Microarray Technology for Molecular Applications: Investigation of Cross-Contamination between Tissue Samples Obtained from the Same Punching Device. Microarrays. 2015; 4(2):188-195. https://doi.org/10.3390/microarrays4020188
Chicago/Turabian StyleVassella, Erik, José A. Galván, and Inti Zlobec. 2015. "Tissue Microarray Technology for Molecular Applications: Investigation of Cross-Contamination between Tissue Samples Obtained from the Same Punching Device" Microarrays 4, no. 2: 188-195. https://doi.org/10.3390/microarrays4020188
APA StyleVassella, E., Galván, J. A., & Zlobec, I. (2015). Tissue Microarray Technology for Molecular Applications: Investigation of Cross-Contamination between Tissue Samples Obtained from the Same Punching Device. Microarrays, 4(2), 188-195. https://doi.org/10.3390/microarrays4020188




