From High-Throughput Microarray-Based Screening to Clinical Application: The Development of a Second Generation Multigene Test for Breast Cancer Prognosis
Abstract
:1. Background—Establishment and Clinical Validation of First Generation Multigene Tests for Breast Cancer Patients
2. Important Aspects for the Establishment and Clinical Validation of Novel Second Generation Multigene Tests
2.1. Biomarker and Molecular Subtypes
2.2. ER+/HER2− Breast Cancer is a Chronic Disease—The Importance of Predicting Late Metastases
2.3. Pitfalls in Study Design—The Importance of Unique Clinical Characteristics and Treatment Strategies in the Training and Validation Phase
2.4. Additional Prognostic Information—Clinical and Molecular Parameters
2.5. Decentral Testing—The Importance of Analytic Validity of Tests and External Proficiency Testing
3. The Establishment of EndoPredict—A Second Generation Multigene Test
3.1. Relevant Patient Group in Training and Validation
3.2. Gene Selection and Algorithm Design
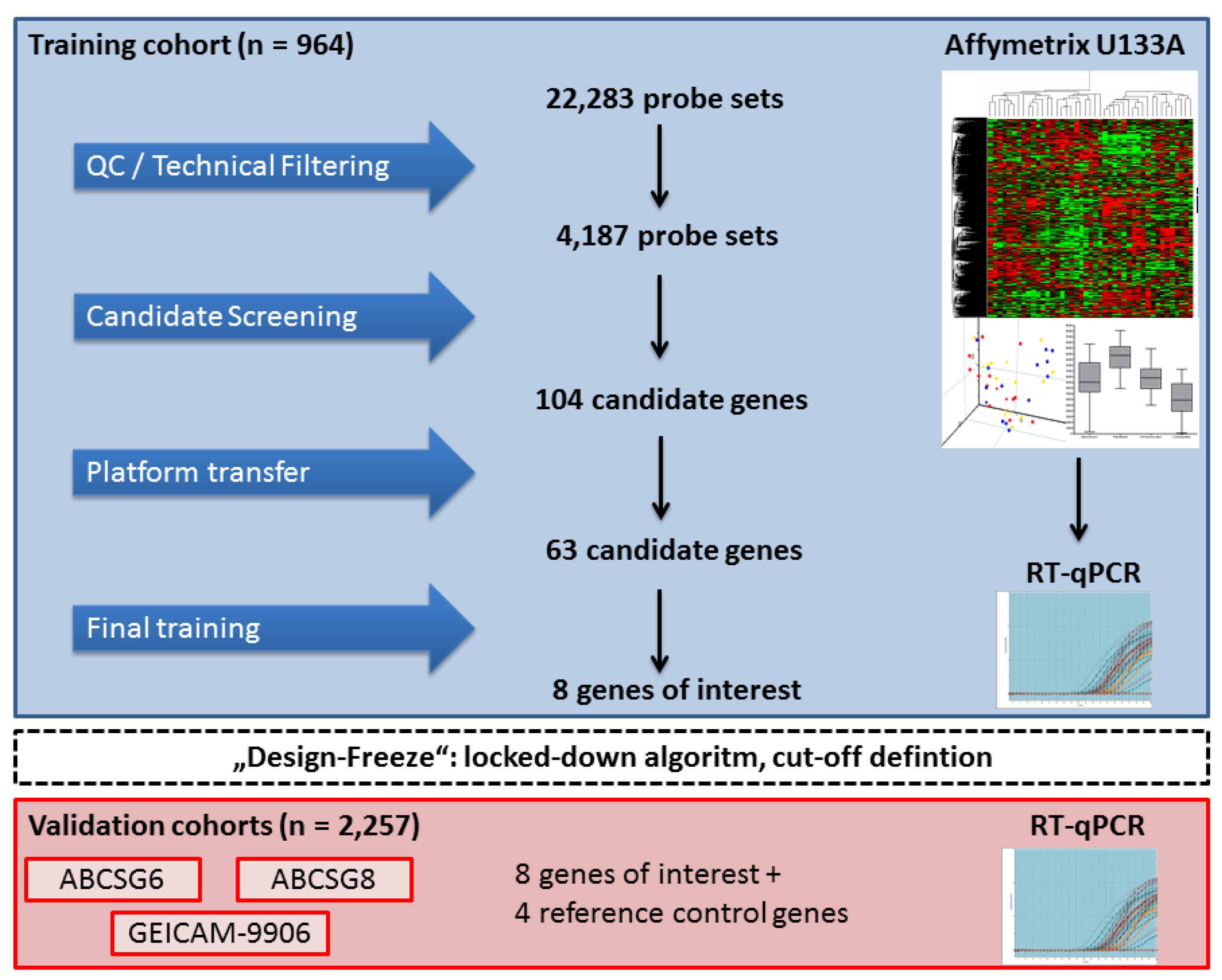
3.3. Independent Clinical Validation of the EndoPredict Test

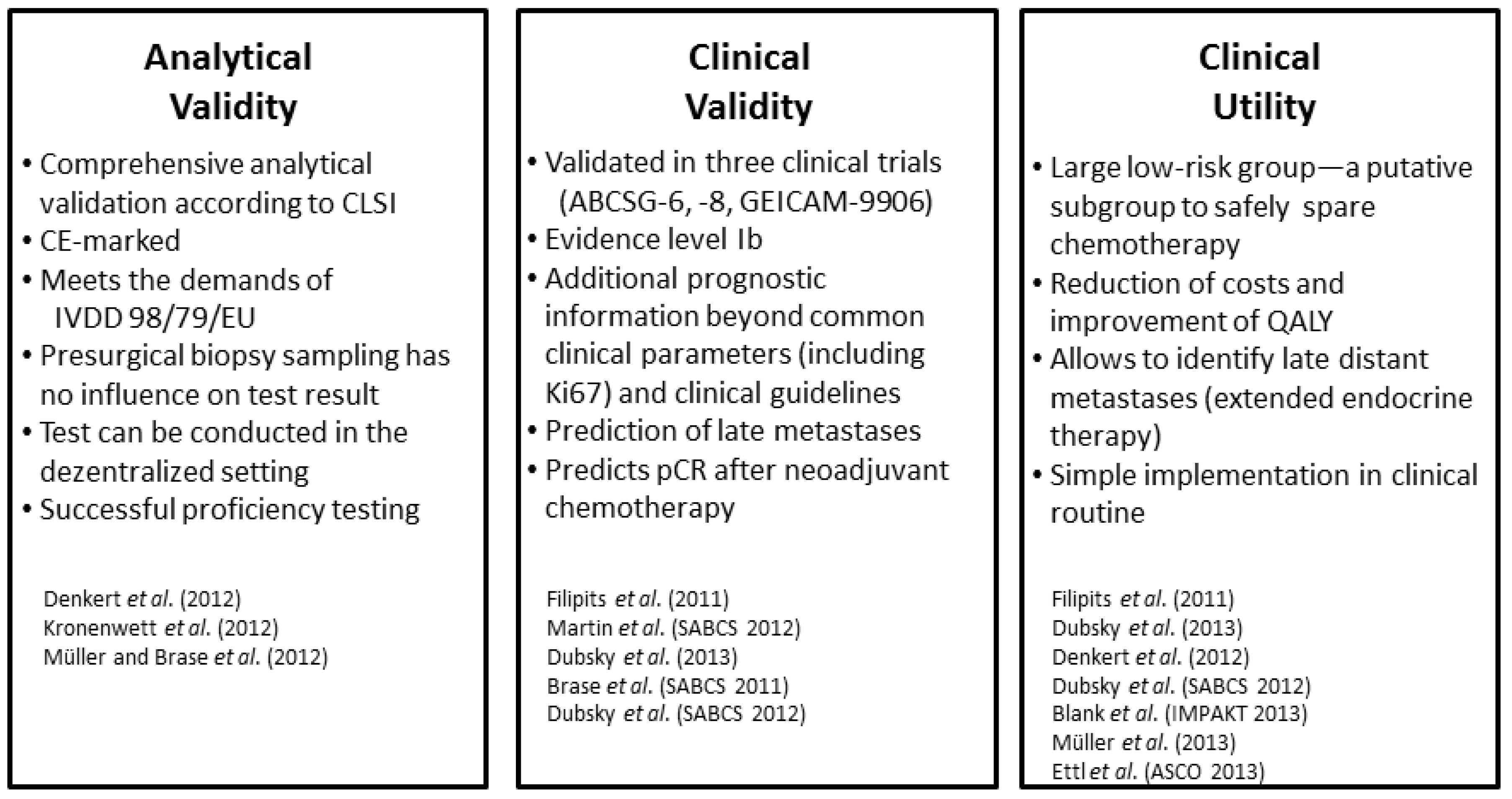
3.4. Analytical Validation and Proficiency Testing
3.5. Clinical Utility—Comparison to Clinical Guidelines, Decision Impact and Health Economics
4. EndoPredict—Ready for Prime Time?
5. Perspectives
Acknowledgements
Conflicts of Interest
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA. Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef]
- Olivotto, I.A.; Bajdik, C.D.; Ravdin, P.M.; Speers, C.H.; Coldman, A.J.; Norris, B.D.; Davis, G.J.; Chia, S.K.; Gelmon, K.A. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J. Clin. Oncol. 2005, 23, 2716–2725. [Google Scholar]
- Galea, M.H.; Blamey, R.W.; Elston, C.E.; Ellis, I.O. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res. Treat. 1992, 22, 207–219. [Google Scholar] [CrossRef]
- D’Eredita, G.; Giardina, C.; Martellotta, M.; Natale, T.; Ferrarese, F. Prognostic factors in breast cancer: The predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur. J. Cancer 2001, 37, 591–596. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Ingle, J.N.; Gelber, R.D.; Coates, A.S.; Thurlimann, B.; Senn, H.J. Thresholds for therapies: Highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann. Oncol. 2009, 20, 1319–1329. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Rouzier, R.; Perou, C.M.; Symmans, W.F.; Ibrahim, N.; Cristofanilli, M.; Anderson, K.; Hess, K.R.; Stec, J.; Ayers, M.; Wagner, P.; et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005, 11, 5678–5685. [Google Scholar] [CrossRef]
- Gluck, S.; de Snoo, F.; Peeters, J.; Stork-Sloots, L.; Somlo, G. Molecular subtyping of early-stage breast cancer identifies a group of patients who do not benefit from neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2013, 139, 759–767. [Google Scholar] [CrossRef]
- Van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef]
- Buyse, M.; Loi, S.; van’t Veer, L.; Viale, G.; Delorenzi, M.; Glas, A.M.; d’Assignies, M.S.; Bergh, J.; Lidereau, R.; Ellis, P.; et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl. Cancer Inst. 2006, 98, 1183–1192. [Google Scholar] [CrossRef]
- Mook, S.; Schmidt, M.K.; Weigelt, B.; Kreike, B.; Eekhout, I.; van de Vijver, M.J.; Glas, A.M.; Floore, A.; Rutgers, E.J.; van’t Veer, L.J. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann. Oncol. 2010, 21, 717–722. [Google Scholar] [CrossRef]
- Wittner, B.S.; Sgroi, D.C.; Ryan, P.D.; Bruinsma, T.J.; Glas, A.M.; Male, A.; Dahiya, S.; Habin, K.; Bernards, R.; Haber, D.A.; et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin. Cancer Res. 2008, 14, 2988–2993. [Google Scholar] [CrossRef]
- Glas, A.M.; Floore, A.; Delahaye, L.J.; Witteveen, A.T.; Pover, R.C.; Bakx, N.; Lahti-Domenici, J.S.; Bruinsma, T.J.; Warmoes, M.O.; Bernards, R.; et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 2006, 7, 278. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef]
- Albain, K.S.; Barlow, W.E.; Shak, S.; Hortobagyi, G.N.; Livingston, R.B.; Yeh, I.T.; Ravdin, P.; Bugarini, R.; Baehner, F.L.; Davidson, N.E.; et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010, 11, 55–65. [Google Scholar] [CrossRef]
- Dowsett, M.; Cuzick, J.; Wale, C.; Forbes, J.; Mallon, E.A.; Salter, J.; Quinn, E.; Dunbier, A.; Baum, M.; Buzdar, A.; et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J. Clin. Oncol. 2010, 28, 1829–1834. [Google Scholar] [CrossRef]
- Cuzick, J.; Dowsett, M.; Pineda, S.; Wale, C.; Salter, J.; Quinn, E.; Zabaglo, L.; Mallon, E.; Green, A.R.; Ellis, I.O.; et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J. Clin. Oncol. 2011, 29, 4273–4278. [Google Scholar] [CrossRef]
- Klang, S.H.; Hammerman, A.; Liebermann, N.; Efrat, N.; Doberne, J.; Hornberger, J. Economic implications of 21-gene breast cancer risk assay from the perspective of an Israeli-managed health-care organization. Value Health 2010, 13, 381–387. [Google Scholar] [CrossRef]
- Partin, J.F.; Mamounas, E.P. Impact of the 21-gene recurrence score assay compared with standard clinicopathologic guidelines in adjuvant therapy selection for node-negative, estrogen receptor-positive breast cancer. Ann. Surg. Oncol. 2011, 18, 3399–3406. [Google Scholar] [CrossRef]
- Bueno-de-Mesquita, J.M.; van Harten, W.H.; Retel, V.P.; van’t Veer, L.J.; van Dam, F.S.; Karsenberg, K.; Douma, K.F.; van Tinteren, H.; Peterse, J.L.; Wesseling, J.; et al. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: A prospective community-based feasibility study (RASTER). Lancet Oncol. 2007, 8, 1079–1087. [Google Scholar] [CrossRef]
- Geffen, D.B.; Amir, N.; Sion-Vardy, N.; Ariad, S.; Kachko, L.; Bayme, M.; Delgado, B.; Dyomin, V.; Argov, S.; Koretz, M. Stage I breast cancer in a regional oncology practice in Israel 2002–2006: Clinicopathologic features, risk estimation and planned therapy of 328 consecutive patients. Breast 2009, 18, 316–321. [Google Scholar] [CrossRef]
- Asad, J.; Jacobson, A.F.; Estabrook, A.; Smith, S.R.; Boolbol, S.K.; Feldman, S.M.; Osborne, M.P.; Boachie-Adjei, K.; Twardzik, W.; Tartter, P.I. Does oncotype DX recurrence score affect the management of patients with early-stage breast cancer? Am. J. Surg. 2008, 196, 527–529. [Google Scholar] [CrossRef]
- Drukker, C.A.; Bueno-de-Mesquita, J.M.; Retel, V.P.; van Harten, W.H.; van Tinteren, H.; Wesseling, J.; Roumen, R.M.; Knauer, M.; van’t Veer, L.J.; Sonke, G.S.; et al. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int. J. Cancer 2013, 133, 929–936. [Google Scholar] [CrossRef]
- Bogaerts, J.; Cardoso, F.; Buyse, M.; Braga, S.; Loi, S.; Harrison, J.A.; Bines, J.; Mook, S.; Decker, N.; Ravdin, P.; et al. Gene signature evaluation as a prognostic tool: Challenges in the design of the MINDACT trial. Nat. Clin. Pract. Oncol. 2006, 3, 540–551. [Google Scholar]
- Cardoso, F.; van’t Veer, L.; Rutgers, E.; Loi, S.; Mook, S.; Piccart-Gebhart, M.J. Clinical application of the 70-gene profile: the MINDACT trial. J. Clin. Oncol. 2008, 26, 729–735. [Google Scholar] [CrossRef]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP working group: Can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet. Med. 2009, 11, 66–73. [CrossRef]
- Fisher, B.; Dignam, J.; Bryant, J.; Wolmark, N. Five versus more than five years of tamoxifen for lymph node-negative breast cancer: Updated findings from the National Surgical Adjuvant Breast and Bowel Project B-14 randomized trial. J. Natl. Cancer Inst. 2001, 93, 684–690. [Google Scholar] [CrossRef]
- Fisher, B.; Jeong, J.H.; Bryant, J.; Anderson, S.; Dignam, J.; Fisher, E.R.; Wolmark, N. Treatment of lymph-node-negative, oestrogen-receptor-positive breast cancer: Long-term findings from National Surgical Adjuvant Breast and Bowel Project randomised clinical trials. Lancet 2004, 364, 858–868. [Google Scholar] [CrossRef]
- Peto, R.; Davies, C.; Godwin, J.; Gray, R.; Pan, H.C.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Taylor, C.; et al. Comparisons between different polychemotherapy regimens for early breast cancer: Meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 2012, 379, 432–444. [Google Scholar] [CrossRef]
- Berry, D.A.; Cirrincione, C.; Henderson, I.C.; Citron, M.L.; Budman, D.R.; Goldstein, L.J.; Martino, S.; Perez, E.A.; Muss, H.B.; Norton, L.; et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 2006, 295, 1658–1667. [Google Scholar] [CrossRef]
- Milburn, M.; Rosman, M.; Mylander, C.; Tafra, L. Is oncotype DX recurrence score (RS) of prognostic value once HER2-positive and low-ER expression patients are removed? Breast J. 2013, 19, 357–364. [Google Scholar] [CrossRef]
- Desmedt, C.; Haibe-Kains, B.; Wirapati, P.; Buyse, M.; Larsimont, D.; Bontempi, G.; Delorenzi, M.; Piccart, M.; Sotiriou, C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 2008, 14, 5158–5165. [Google Scholar] [CrossRef]
- Schmidt, M.; Bohm, D.; von Torne, C.; Steiner, E.; Puhl, A.; Pilch, H.; Lehr, H.A.; Hengstler, J.G.; Kolbl, H.; Gehrmann, M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008, 68, 5405–5413. [Google Scholar] [CrossRef]
- Schmidt, M.; Hellwig, B.; Hammad, S.; Othman, A.; Lohr, M.; Chen, Z.; Boehm, D.; Gebhard, S.; Petry, I.; Lebrecht, A.; et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin к C as a compatible prognostic marker in human solid tumors. Clin. Cancer Res. 2012, 18, 2695–2703. [Google Scholar] [CrossRef]
- Schmidt, M.; Hengstler, J.G.; von Torne, C.; Koelbl, H.; Gehrmann, M.C. Coordinates in the universe of node-negative breast cancer revisited. Cancer Res. 2009, 69, 2695–2698. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Gomez, S.; Arenas, A.; El-Ashry, D.; Schmidt, M.; Gehrmann, M.; Caldas, C. Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules. BMC Cancer 2010, 10, 604. [Google Scholar] [CrossRef]
- Bianchini, G.; Qi, Y.; Alvarez, R.H.; Iwamoto, T.; Coutant, C.; Ibrahim, N.K.; Valero, V.; Cristofanilli, M.; Green, M.C.; Radvanyi, L.; et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J. Clin. Oncol. 2010, 28, 4316–4323. [Google Scholar] [CrossRef]
- Esserman, L.J.; Moore, D.H.; Tsing, P.J.; Chu, P.W.; Yau, C.; Ozanne, E.; Chung, R.E.; Tandon, V.J.; Park, J.W.; Baehner, F.L.; et al. Biologic markers determine both the risk and the timing of recurrence in breast cancer. Breast Cancer Res. Treat. 2011, 129, 607–616. [Google Scholar] [CrossRef]
- Jatoi, I.; Anderson, W.F.; Jeong, J.H.; Redmond, C.K. Breast cancer adjuvant therapy: Time to consider its time-dependent effects. J. Clin. Oncol. 2011, 29, 2301–2304. [Google Scholar] [CrossRef]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Alencar, V.H.; Badran, A.; Bonfill, X.; et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013, 381, 805–816. [Google Scholar] [CrossRef]
- Goss, P.E. Letrozole in the extended adjuvant setting: MA.17. Breast Cancer Res. Treat. 2007, 105 Suppl 1, 45–53. [Google Scholar] [CrossRef]
- Goss, P.E.; Ingle, J.N.; Martino, S.; Robert, N.J.; Muss, H.B.; Livingston, R.B.; Davidson, N.E.; Perez, E.A.; Chavarri-Guerra, Y.; Cameron, D.A.; et al. Impact of premenopausal status at breast cancer diagnosis in women entered on the placebo-controlled NCIC CTG MA17 trial of extended adjuvant letrozole. Ann. Oncol. 2013, 24, 355–361. [Google Scholar] [CrossRef]
- Goss, P.E.; Ingle, J.N.; Martino, S.; Robert, N.J.; Muss, H.B.; Piccart, M.J.; Castiglione, M.; Tu, D.; Shepherd, L.E.; Pritchard, K.I.; et al. Randomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: Updated findings from NCIC CTG MA.17. J. Natl. Cancer Inst. 2005, 97, 1262–1271. [Google Scholar] [CrossRef]
- Goss, P.E.; Ingle, J.N.; Martino, S.; Robert, N.J.; Muss, H.B.; Piccart, M.J.; Castiglione, M.; Tu, D.; Shepherd, L.E.; Pritchard, K.I.; et al. Efficacy of letrozole extended adjuvant therapy according to estrogen receptor and progesterone receptor status of the primary tumor: National Cancer Institute of Canada Clinical Trials Group MA.17. J. Clin. Oncol. 2007, 25, 2006–2011. [Google Scholar] [CrossRef]
- Goss, P.E.; Ingle, J.N.; Martino, S.; Robert, N.J.; Muss, H.B.; Piccart, M.J.; Castiglione, M.; Tu, D.; Shepherd, L.E.; Pritchard, K.I.; et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N. Engl. J. Med. 2003, 349, 1793–1802. [Google Scholar] [CrossRef]
- Mamounas, E.P.; Jeong, J.H.; Wickerham, D.L.; Smith, R.E.; Ganz, P.A.; Land, S.R.; Eisen, A.; Fehrenbacher, L.; Farrar, W.B.; Atkins, J.N.; et al. Benefit from exemestane as extended adjuvant therapy after 5 years of adjuvant tamoxifen: Intention-to-treat analysis of the National Surgical Adjuvant Breast And Bowel Project B-33 trial. J. Clin. Oncol. 2008, 26, 1965–1971. [Google Scholar] [CrossRef]
- Jakesz, R.; Greil, R.; Gnant, M.; Schmid, M.; Kwasny, W.; Kubista, E.; Mlineritsch, B.; Tausch, C.; Stierer, M.; Hofbauer, F.; et al. Extended adjuvant therapy with anastrozole among postmenopausal breast cancer patients: Results from the randomized Austrian Breast and Colorectal Cancer Study Group Trial 6a. J. Natl. Cancer Inst. 2007, 99, 1845–1853. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Sestak, I.; Cuzick, J.; Zhang, Y.; Schnabel, C.A.; Erlander, M.G.; Goss, P.E.; Dowsett, M. Comparative performance of breast cancer Index (BCI) vs. oncotype Dx and IHC4 in the prediction of late recurrence in hormonal receptor-positive lymph node-negative breast cancer patients: A TransATAC study. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
- Cheang, M.C.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Varga, Z.; Diebold, J.; Dommann-Scherrer, C.; Frick, H.; Kaup, D.; Noske, A.; Obermann, E.; Ohlschlegel, C.; Padberg, B.; Rakozy, C.; et al. How reliable is Ki-67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss Working Group of Breast- and Gynecopathologists. PLoS One 2012, 7, e37379. [Google Scholar] [CrossRef] [Green Version]
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011, 17, 6012–6020. [Google Scholar] [CrossRef]
- Patterson, T.A.; Lobenhofer, E.K.; Fulmer-Smentek, S.B.; Collins, P.J.; Chu, T.M.; Bao, W.; Fang, H.; Kawasaki, E.S.; Hager, J.; Tikhonova, I.R.; et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 2006, 24, 1140–1150. [Google Scholar] [CrossRef]
- Shi, L.; Reid, L.H.; Jones, W.D.; Shippy, R.; Warrington, J.A.; Baker, S.C.; Collins, P.J.; de Longueville, F.; Kawasaki, E.S.; Lee, K.Y.; et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006, 24, 1151–1161. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Warren, D.; Spencer, F.; Kim, I.F.; Biswal, S.; Frank, B.C.; Gabrielson, E.; Garcia, J.G.; Geoghegan, J.; Germino, G.; et al. Multiple-laboratory comparison of microarray platforms. Nat. Methods 2005, 2, 345–350. [Google Scholar] [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Furness, P.N.; Taub, N.; Assmann, K.J.; Banfi, G.; Cosyns, J.P.; Dorman, A.M.; Hill, C.M.; Kapper, S.K.; Waldherr, R.; Laurinavicius, A.; et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am. J. Surg. Pathol. 2003, 27, 805–810. [Google Scholar] [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef]
- Desmedt, C.; Giobbie-Hurder, A.; Neven, P.; Paridaens, R.; Christiaens, M.R.; Smeets, A.; Lallemand, F.; Haibe-Kains, B.; Viale, G.; Gelber, R.D.; et al. The Gene expression Grade Index: A potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1–98 trial. BMC Med. Genom. 2009, 2, 40. [Google Scholar] [CrossRef]
- Loi, S.; Haibe-Kains, B.; Desmedt, C.; Lallemand, F.; Tutt, A.M.; Gillet, C.; Ellis, P.; Harris, A.; Bergh, J.; Foekens, J.A.; et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 2007, 25, 1239–1246. [Google Scholar] [CrossRef]
- Bohmann, K.; Hennig, G.; Rogel, U.; Poremba, C.; Mueller, B.M.; Fritz, P.; Stoerkel, S.; Schaefer, K.L. RNA extraction from archival formalin-fixed paraffin-embedded tissue: A comparison of manual, semiautomated, and fully automated purification methods. Clin. Chem. 2009, 55, 1719–1727. [Google Scholar] [CrossRef]
- Hennig, G.; Gehrmann, M.; Stropp, U.; Brauch, H.; Fritz, P.; Eichelbaum, M.; Schwab, M.; Schroth, W. Automated extraction of DNA and RNA from a single formalin-fixed paraffin-embedded tissue section for analysis of both single-nucleotide polymorphisms and mRNA expression. Clin. Chem. 2010, 56, 1845–1853. [Google Scholar] [CrossRef]
- Muller, B.M.; Kronenwett, R.; Hennig, G.; Euting, H.; Weber, K.; Bohmann, K.; Weichert, W.; Altmann, G.; Roth, C.; Winzer, K.J.; et al. Quantitative determination of estrogen receptor, progesterone receptor, and HER2 mRNA in formalin-fixed paraffin-embedded tissue—A new option for predictive biomarker assessment in breast cancer. Diagn. Mol. Pathol. 2011, 20, 1–10. [Google Scholar] [CrossRef]
- Dubsky, P.; Brase, J.C.; Fisch, K.; Jakesz, R.; Singer, C.F.; Greil, R.; Dietze, O.; Weber, K.E.; Petry, C.; Kronenwett, R.; et al. The EndoPredict score identifies late distant metastases in ER+/HER2− breast cancer patients. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
- Ein-Dor, L.; Kela, I.; Getz, G.; Givol, D.; Domany, E. Outcome signature genes in breast cancer: Is there a unique set? Bioinformatics 2005, 21, 171–178. [Google Scholar] [CrossRef]
- Dubsky, P.C.; Jakesz, R.; Mlineritsch, B.; Postlberger, S.; Samonigg, H.; Kwasny, W.; Tausch, C.; Stoger, H.; Haider, K.; Fitzal, F.; et al. Tamoxifen and anastrozole as a sequencing strategy: A randomized controlled trial in postmenopausal patients with endocrine-responsive early breast cancer from the Austrian Breast and Colorectal Cancer Study Group. J. Clin. Oncol. 2012, 30, 722–728. [Google Scholar] [CrossRef]
- Simon, R.M.; Paik, S.; Hayes, D.F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl. Cancer Inst. 2009, 101, 1446–1452. [Google Scholar] [CrossRef]
- Martin, M.; Brase, J.C.; Ruiz-Borrego, M.; Krappmann, K.; Munarriz, B.; Fisch, K.; Ruiz, A.; Weber, K.E.; Crespo, C.; Petry, C.; et al. Prognostic performance of the EndoPredict score in node-positive chemotherapy-treated ER+/HER2− breast cancer patients: results from the GEICAM/9906 trial. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
- Brase, J.C.; Gehrmann, M.C.; Petry, C.; Weber, K.E.; Schmidt, M.; Kölbl, H.; Schroth, W.; Schwab, M.; Müller, V.; Jänicke, F.; et al. The EndoPredict score is a response predictor for neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer. Cancer Res. 2011, 71. [Google Scholar] [CrossRef]
- Kronenwett, R.; Bohmann, K.; Prinzler, J.; Sinn, B.V.; Haufe, F.; Roth, C.; Averdick, M.; Ropers, T.; Windbergs, C.; Brase, J.C.; et al. Decentral gene expression analysis: Analytical validation of the Endopredict genomic multianalyte breast cancer prognosis test. BMC Cancer 2012, 12, 456. [Google Scholar] [CrossRef]
- Denkert, C.; Kronenwett, R.; Schlake, W.; Bohmann, K.; Penzel, R.; Weber, K.E.; Hofler, H.; Lehmann, U.; Schirmacher, P.; Specht, K.; et al. Decentral gene expression analysis for ER+/HER2− breast cancer: Results of a proficiency testing program for the EndoPredict assay. Virchows Arch. 2012, 460, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Noske, A.; Loibl, S.; Darb-Esfahani, S.; Roller, M.; Kronenwett, R.; Muller, B.M.; Steffen, J.; von Toerne, C.; Wirtz, R.; Baumann, I.; et al. Comparison of different approaches for assessment of HER2 expression on protein and mRNA level: Prediction of chemotherapy response in the neoadjuvant GeparTrio trial (NCT00544765). Breast Cancer Res. Treat. 2011, 126, 109–117. [Google Scholar] [CrossRef]
- Loibl, S.; Muller, B.M.; von Minckwitz, G.; Schwabe, M.; Roller, M.; Darb-Esfahani, S.; Ataseven, B.; du Bois, A.; Fissler-Eckhoff, A.; Gerber, B.; et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2011, 130, 477–487. [Google Scholar] [CrossRef]
- Muller, B.M.; Brase, J.C.; Haufe, F.; Weber, K.E.; Budzies, J.; Petry, C.; Prinzler, J.; Kronenwett, R.; Dietel, M.; Denkert, C. Comparison of the RNA-based EndoPredict multigene test between core biopsies and corresponding surgical breast cancer sections. J. Clin. Pathol. 2012, 65, 660–662. [Google Scholar] [CrossRef]
- Wöckel, A.; Kreienberg, R. First revision of the German S3 guideline ‘diagnosis, Therapy, and Follow-Up of Breast Cancer’. Breast Care (Basel) 2008, 3, 82–86. [Google Scholar] [CrossRef]
- Carlson, R.W.; Brown, E.; Burstein, H.J.; Gradishar, W.J.; Hudis, C.A.; Loprinzi, C.; Mamounas, E.P.; Perez, E.A.; Pritchard, K.; Ravdin, P.; et al. NCCN task force report: Adjuvant therapy for breast cancer. J. Natl. Compr. Canc. Netw. 2006, 4 Suppl 1, S1–S26. [Google Scholar]
- Dubsky, P.; Filipits, M.; Jakesz, R.; Rudas, M.; Singer, C.F.; Greil, R.; Dietze, O.; Luisser, I.; Klug, E.; Sedivy, R.; et al. EndoPredict improves the prognostic classification derived from common clinical guidelines in ER-positive, HER2-negative early breast cancer. Ann. Oncol. 2013, 24, 640–647. [Google Scholar] [CrossRef]
- Muller, B.M.; Keil, E.; Lehmann, A.; Winzer, K.J.; Richter-Ehrenstein, C.; Prinzler, J.; Bangemann, N.; Reles, A.; Stadie, S.; Schoenegg, W.; et al. The EndoPredict gene-expression assay in clinical practice—Performance and impact on clinical decisions. PLoS One 2013, 8, e68252. [Google Scholar] [CrossRef]
- Ettl, J.; Große Lackmann, K.; Hapfelmeier, A.; Klein, E.; Paepke, S.; Petry, C.; Specht, K.; Höfler, H.; Kiechle, M. Prospective Comparison of uPA/PAI-1 and Endopredict-Clin Score in ER-Positive, HER2-Negative Breast Cancer: Impact on Risk Stratification and Treatment Decisions. In Proceeding of 2013 ASCO Annual Meeting, Chicago, IL, USA, 31 May–4 June 2013.
- Blank, P.; Schwenkglenks, M.; Dubsky, P.; Filipits, M.; Gutzwiller, F.; Lux, M.P.; Brase, J.C.; Kronenwett, R.; Szucs, T.D.; Gnant, M. Health economic analysis of guideline and gene expression signature-based risk stratification of distant recurrence in early breast cancer patients. Ann. Oncol. 2013, 24. [Google Scholar] [CrossRef]
- Weigelt, B.; Reis-Filho, J.S.; Swanton, C. Genomic analyses to select patients for adjuvant chemotherapy: Trials and tribulations. Ann. Oncol. 2012, 23 Suppl 10, x211–x218. [Google Scholar]
- Nielsen, T.O.; Parker, J.S.; Leung, S.; Voduc, D.; Ebbert, M.; Vickery, T.; Davies, S.R.; Snider, J.; Stijleman, I.J.; Reed, J.; et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 2010, 16, 5222–5232. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Jankowitz, R.C.; Cooper, K.; Erlander, M.G.; Ma, X.J.; Kesty, N.C.; Li, H.; Chivukula, M.; Brufsky, A. Prognostic utility of the breast cancer index and comparison to Adjuvant! Online in a clinical case series of early breast cancer. Breast Cancer Res. 2011, 13, R98. [Google Scholar] [CrossRef]
- Jerevall, P.L.; Ma, X.J.; Li, H.; Salunga, R.; Kesty, N.C.; Erlander, M.G.; Sgroi, D.C.; Holmlund, B.; Skoog, L.; Fornander, T.; et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br. J. Cancer 2011, 104, 1762–1769. [Google Scholar] [CrossRef]
- Varga, Z.; Sinn, P.; Fritzsche, F.; von Hochstetter, A.; Noske, A.; Schraml, P.; Tausch, C.; Trojan, A.; Moch, H. Comparison of EndoPredict and oncotype DX test results in hormone receptor positive invasive breast cancer. PLoS One 2013, 8, e58483. [Google Scholar] [CrossRef] [Green Version]
- Dowsett, M.; Sestak, I.; Lopez-Knowles, E.; Sidhu, K.; Dunbier, A.K.; Cowens, J.W.; Ferree, S.; Storhoff, J.; Schaper, C.; Cuzick, J. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J. Clin. Oncol. 2013. [Google Scholar] [CrossRef]
- Mamounas, E.P.; Tang, G.; Paik, S.; Baehner, F.L.; Liu, Q.; Jeong, J.H.; Kim, S.R.; Butler, S.M.; Jamshidian, F.; Cherbavaz, D.B.; et al. Association between the 21-gene recurrence score (RS) and benefit from adjuvant paclitaxel (Pac) in node-positive (N+), ER-positive breast cancer patients (pts): Results from NSABP B-28. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Brase, J.C.; Kronenwett, R.; Petry, C.; Denkert, C.; Schmidt, M. From High-Throughput Microarray-Based Screening to Clinical Application: The Development of a Second Generation Multigene Test for Breast Cancer Prognosis. Microarrays 2013, 2, 243-264. https://doi.org/10.3390/microarrays2030243
Brase JC, Kronenwett R, Petry C, Denkert C, Schmidt M. From High-Throughput Microarray-Based Screening to Clinical Application: The Development of a Second Generation Multigene Test for Breast Cancer Prognosis. Microarrays. 2013; 2(3):243-264. https://doi.org/10.3390/microarrays2030243
Chicago/Turabian StyleBrase, Jan C., Ralf Kronenwett, Christoph Petry, Carsten Denkert, and Marcus Schmidt. 2013. "From High-Throughput Microarray-Based Screening to Clinical Application: The Development of a Second Generation Multigene Test for Breast Cancer Prognosis" Microarrays 2, no. 3: 243-264. https://doi.org/10.3390/microarrays2030243




