Three Soil Bacterial Communities from an Archaeological Excavation Site of an Ancient Coal Mine near Bennstedt (Germany) Characterized by 16S r-RNA Sequencing
Abstract
:1. Introduction
2. Experimental
2.1. Excavation Site and Soil Samples
2.2. DNA Extraction, Amplification, and Labelling
2.3. Processing of NGS Data
2.4. Data Analyses
3. Results and Discussion
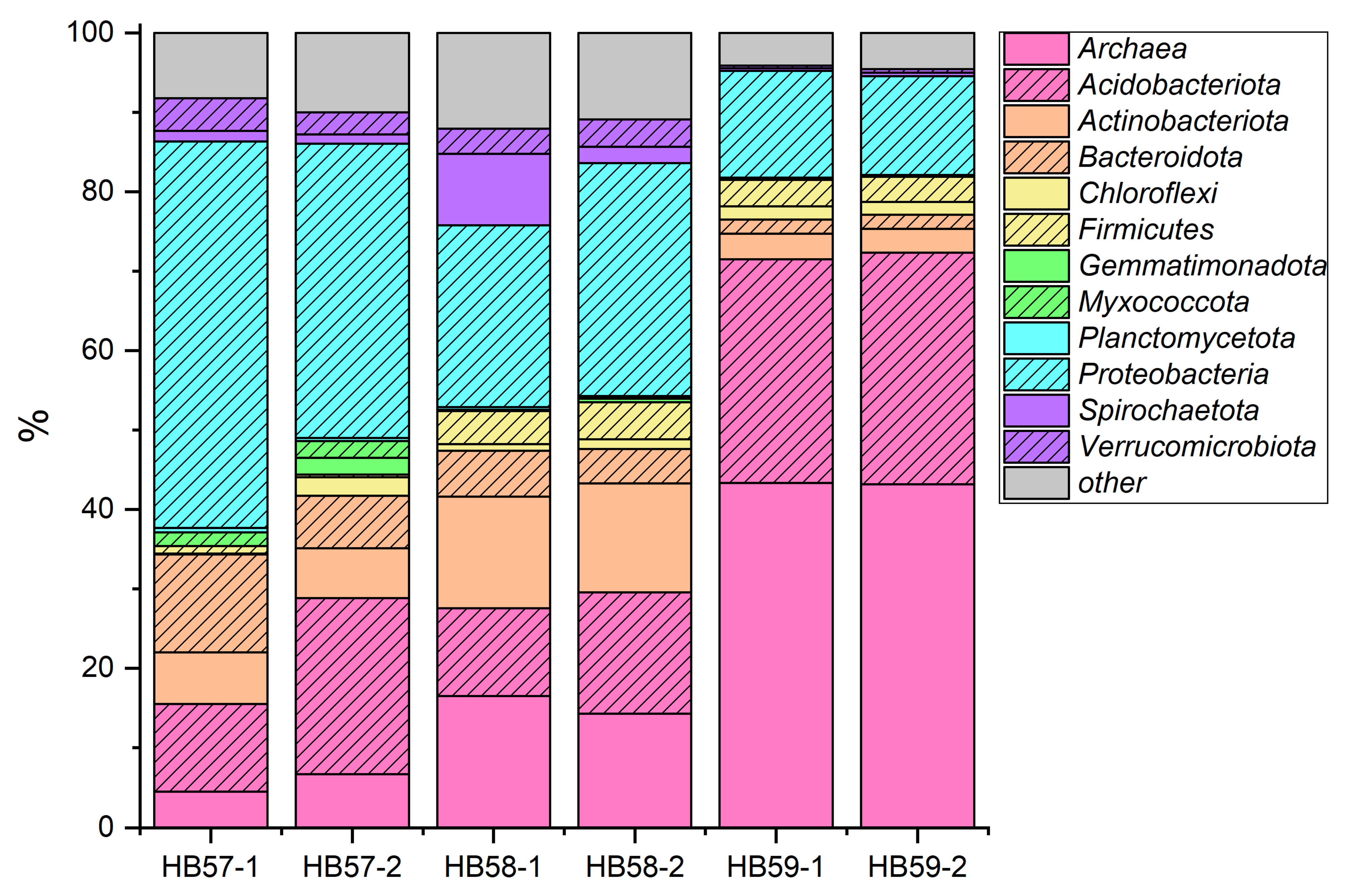
3.1. Comparison of Sampling Sites According to the Bacterial Soil Community at Phylum Level
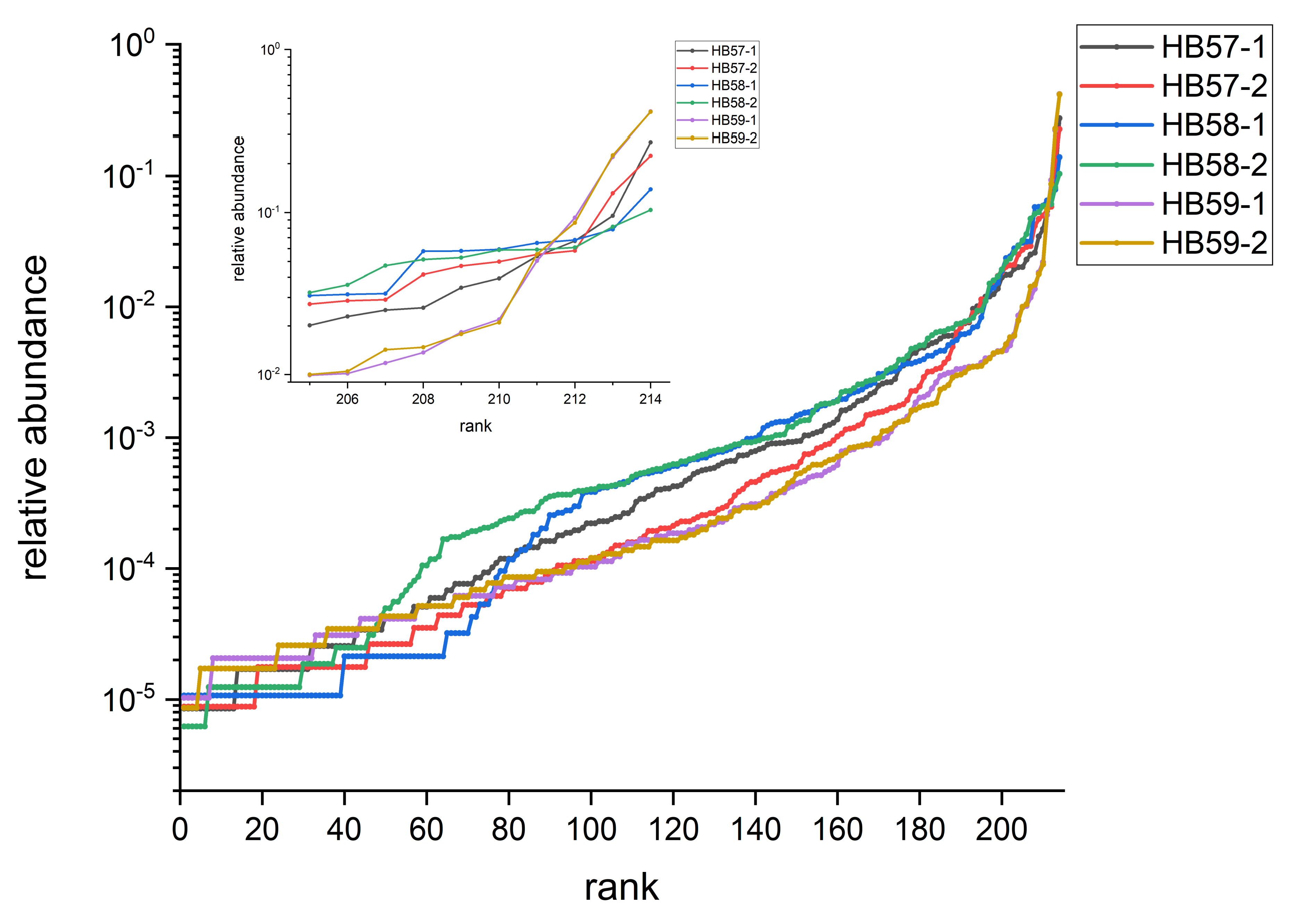
3.2. Rank Profiles of OTU Abundances
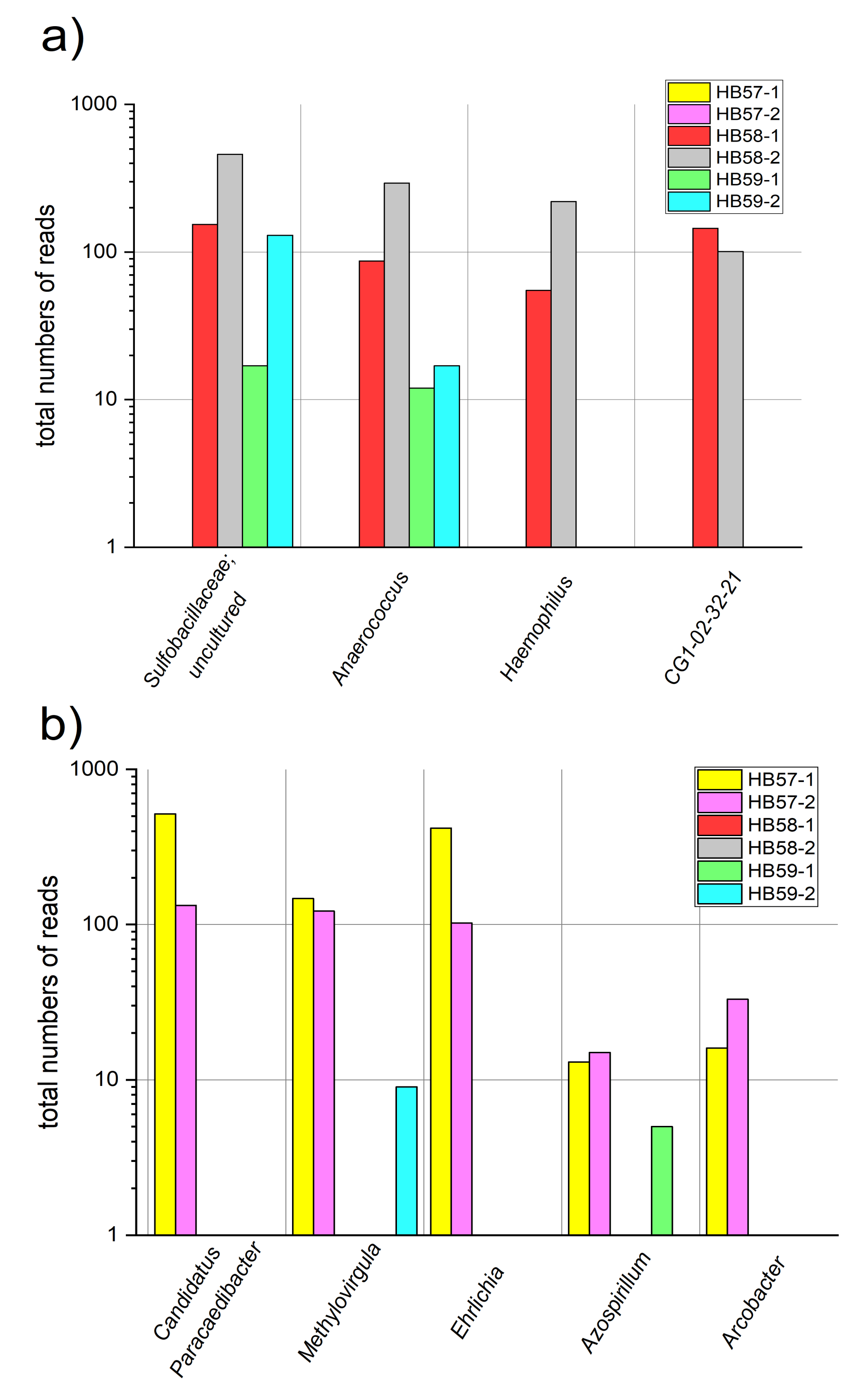
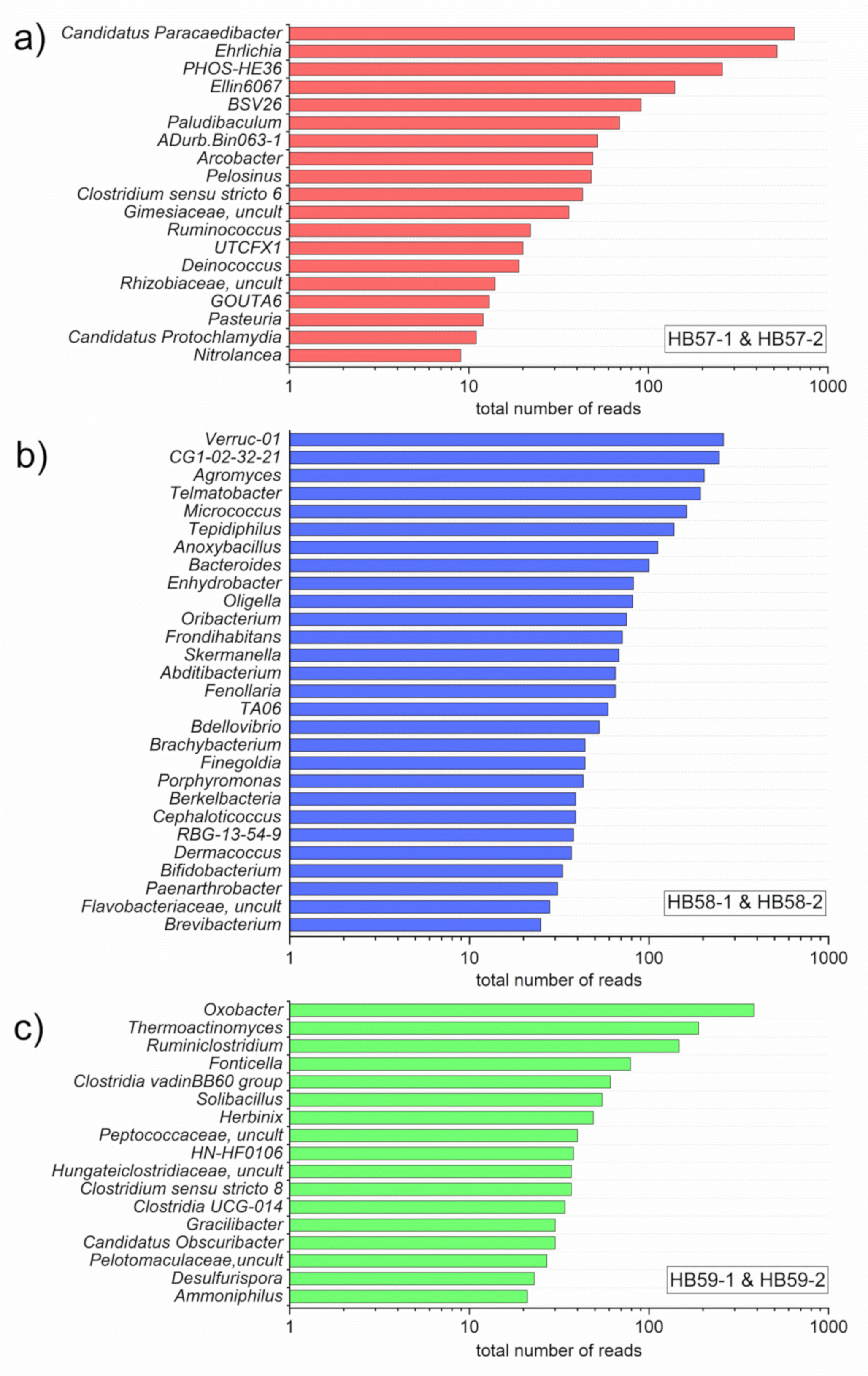
3.3. Comparison of Sampling Sites by Special OTUs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torsvik, V.; Sorheim, R.; Gokskoyr, J. Total bacterial diversity in soil and sediment—A review. J. Ind. Microbiol. 1996, 17, 170–178. [Google Scholar] [CrossRef]
- Romanowicz, K.J.; Freedman, Z.B.; Upchurch, R.A.; Argiroff, W.A.; Zak, D.R. Active microorganisms in forest soils differ from the total community yet are shaped by the same environmental factors: The influence of pH and soil moisture. FEMS Microbiol. Ecol. 2016, 92, 149. [Google Scholar] [CrossRef] [PubMed]
- Rathore, P.; Joy, S.S.; Yadav, R.; Ramakrishna, W. Co-occurrence and patterns of phosphate solubilizing, salt and metal tolerant and antibiotic-resistant bacteria in diverse soils. 3 Biotech 2021, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Pecher, W.T.; Al Madadha, M.E.; DasSarma, P.; Ekulona, F.; Schott, E.J.; Crowe, K.; Gut, B.S.; DasSarma, S. Effects of road salt on microbial communities: Halophiles as biomarkers of road salt pollution. PLoS ONE 2019, 14, e0221355. [Google Scholar] [CrossRef] [PubMed]
- Haferburg, G.; Kothe, E. Metallomics: Lessons for metalliferous soil remediation. Appl. Microbiol. Biotechnol. 2010, 87, 1271–1280. [Google Scholar] [CrossRef]
- Schippers, A.; Jozsa, P.; Sand, W. Sulfur chemistry in bacterial leaching of pyrite. Appl. Environ. Microbiol. 1996, 62, 3424–3431. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hua, Z.-S.; Chen, L.-X.; Kuang, J.-L.; Li, S.-J.; Shu, W.-S.; Huang, L.-N. Correlating Microbial Diversity Patterns with Geochemistry in an Extreme and Heterogeneous Environment of Mine Tailings. Appl. Environ. Microbiol. 2014, 80, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Méndez-García, C.; Mesa, V.; Sprenger, R.R.; Richter, M.; Diez, M.S.; Solano, J.; Bargiela, R.; Golyshina, O.V.; Manteca, Á.; Ramos, J.L.; et al. Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage. ISME J. 2014, 8, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- Thavamani, P.; Samkumar, R.A.; Sathees, V.; Subashchandrabose, S.R.; Ramadass, K.; Naidu, R.; Venkateswarlu, K.; Megharaj, M. Microbes from mined sites: Harnessing their potential for reclamation of derelicted mine sites. Environ. Pollut. 2017, 230, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Mesa, V.; Gallego, J.L.R.; Gonzáles-Gil, R.; Lauga, B.; Sánchez, J.; Méndez-Garcia, C.; Peláez, A.I. Bacterial, archaeal, and eukaryotic diversity across distinct microhabits in an acid mine drainage. Front. Microbiol. 2018, 8, 1756. [Google Scholar] [CrossRef]
- Buta, M.; Blaga, G.; Paulette, L.; Păcurar, I.; Roșca, S.; Borsai, O.; Grecu, F.; Sînziana, P.E.; Negrușier, C. Soil Reclamation of Abandoned Mine Lands by Revegetation in Northwestern Part of Transylvania: A 40-Year Retrospective Study. Sustainability 2019, 11, 3393. [Google Scholar] [CrossRef]
- Olías, M.; Nieto, J.M. Background Conditions and Mining Pollution throughout History in the Río Tinto (SW Spain). Environments 2015, 2, 295–316. [Google Scholar] [CrossRef]
- Mourinha, C.; Palma, P.; Alexandre, C.; Cruz, N.; Rodrigues, S.M.; Alvarenga, P. Potentially toxic elements contamination of soils affected by mining activities in the Portugese sector of the Iberian pyrite belt and optional remediation actions, a review. Environments 2022, 9, 11. [Google Scholar] [CrossRef]
- Wolińska, A.; Włodarczyk, K.; Kuźniar, A.; Marzec-Grządziel, A.; Grządziel, J.; Gałązka, A.; Uzarowicz, Ł. Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy 2020, 10, 1795. [Google Scholar] [CrossRef]
- Bartelt-Ryser, J.; Joshi, J.; Schmid, B.; Brandl, H.; Balser, T. Soil feedbacks of plant diversity on soil microbial communities and subsequent plant growth. Perspect. Plant Ecol. Evol. Syst. 2005, 7, 27–49. [Google Scholar] [CrossRef]
- Jones, S.E.; Lennon, J.T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA 2010, 107, 5881–5886. [Google Scholar] [CrossRef] [PubMed]
- Aanderud, Z.T.; Jones, S.E.; Fierer, N.; Lennon, J.T. Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front. Microbiol. 2015, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria Phylum Sequences in Uranium-Contaminated Subsurface Sediments Greatly Expand the Known Diversity within the Phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Epstein, S.; D’Onofrio, A.; Ling, L. L Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010, 63, 468–476. [Google Scholar] [CrossRef]
- Luo, Z.; Ma, J.; Chen, F.; Li, X.; Hou, H.; Zhang, S. Cracks Reinforce the Interactions among Soil Bacterial Communities in the Coal Mining Area of Loess Plateau, China. Int. J. Environ. Res. Public Health 2019, 16, 4892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, J.M.; Kalensee, F.; Günther, P.M.; Schüler, T.; Cao, J. The Local Ecological Memory of Soil: Majority and Minority Components of Bacterial Communities in Prehistorical Urns from Schöps (Germany). Int. J. Environ. Res. 2018, 12, 575–584. [Google Scholar] [CrossRef]
- Warinner, C.; Herbig, A.; Mann, A.; Yates, J.A.F.; Weiß, C.L.; Burbano, H.A.; Orlando, L.; Krause, J. A Robust Framework for Microbial Archaeology. Annu. Rev. Genom. Hum. Genet. 2017, 18, 321–356. [Google Scholar] [CrossRef]
- Philips, A.; Stolarek, I.; Kuczkowska, B.; Juras, A.; Handschuh, L.; Piontek, J.; Kozlowski, P.; Figlerowicz, M. Comprehensive analysis of microorganisms accompanying human archaeological remains. GigaScience 2017, 6, 1–13. [Google Scholar] [CrossRef]
- Lenehan, C.E.; Tobe, S.S.; Smith, R.J.; Popelka-Filcoff, R.S. Microbial composition analyses by 16S rRNA sequencing: A proof of concept approach to provenance determination of archaeological ochre. PLoS ONE 2017, 12, e0185252. [Google Scholar] [CrossRef]
- Xu, J.; Wei, Y.; Jia, H.; Xiao, L.; Gong, D. A new perspective on studying burial environment before archaeological excavation: Analysing bacterial community distribution by high-throughput sequencing. Sci. Rep. 2017, 7, 41691. [Google Scholar] [CrossRef] [PubMed]
- Chernysheva, E.; Korobov, D.; Borisov, A. Thermophilic microorganisms in arable land around medieval archaeological sites in Northern Caucasus, Russia: Novel evidence of past manuring practices. Geoarchaeology 2017, 32, 494–501. [Google Scholar] [CrossRef]
- Margesin, R.; Siles, J.A.; Cajthaml, T.; Öhlinger, B.; Kistler, E. Microbiology Meets Archaeology: Soil Microbial Communities Reveal Different Human Activities at Archaic Monte Iato (Sixth Century BC). Microb. Ecol. 2016, 73, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Sabin, S.; Yeh, H.-Y.; Pluskowski, A.; Clamer, C.; Mitchell, P.D.; Bos, K.I. Estimating molecular preservation of the intestinal microbiome via metagenomics analyses of latrine sediments from two medieval cities. Philos. Trans. R. Soc. B 2020, 375, 0576. [Google Scholar] [CrossRef] [PubMed]
- Köhler, J.M.; Beetz, N.; Günther, P.M.; Möller, F.; Schüler, T.; Cao, J. Microbial community types and signature-like soil bacterial patterns from fortified prehistoric hills of Thuringia (Germany). Community Ecol. 2020, 21, 107–120. [Google Scholar] [CrossRef]
- Wan, S.; Xia, M.; Tao, J.; Pang, Y.; Yu, F.; Wu, J.; Chen, S. Metagenomics Analysis Reveals the Microbial Communities, Antimicrobial Resistance Gene Diversity and Potential Pathogen Transmission Risk of Two Different Landfills in China. Diversity 2021, 13, 230. [Google Scholar] [CrossRef]
- Wegner, C.-E.; Liesack, W. Unexpected Dominance of Elusive Acidobacteria in Early Industrial Soft Coal Slags. Front. Microbiol. 2017, 8, 1023. [Google Scholar] [CrossRef] [PubMed]
- Köhler, J.M.; Kalensee, F.; Günther, P.M.; Cao, J. Searching for Rare Type Associations in Bacterial Communities from Ancient Copper Mining Areas in The East Harz Region (Germany). Asp. Min. Miner. Sci. 2018, 2, 1–14. [Google Scholar] [CrossRef]
- Köhler, J.M.; Kalensee, F.; Cao, J.; Günther, P.M. Hadesarchaea and other extremophile bacteria from ancient mining areas of the East Harz region (Germany) suggest an ecological long-term memory of soil. SN Appl. Sci. 2019, 1, 839. [Google Scholar] [CrossRef]
- Singer, D.; Herndon, E.; Zemanek, L.; Kortney, C.; Sander, T.; Senko, J.; Perdrial, N. Biogeochemical controls on the potantial for long-term contaminated leaching from soils developing on historical coal mine spoil. Soil Syst. 2021, 5, 3. [Google Scholar] [CrossRef]
- Schumann, P.; Kalensee, F.; Cao, J.; Criscuolo, A.; Clermont, D.; Köhler, J.M.; Meier-Kolthoff, J.P.; Neumann-Schaal, M.; Tindall, B.J.; Pukall, R. Reclassification of halobacterium glacieicola as Occultella glacieicola gen. nov., comb. Nov, of Haloactinobacterium album as Runana alba comb. nov, with an emended description of the genus Ruania, recognition that the genus names Haloactinobacterium and Ruania are heterotopic synonyms and descrption of Occultella aeris sp. nov., a halotolerant isolate from surface soil sampled at an ancient copper smelter. Int. J. Syst. Evol. Microbiol. 2021, 71, 004769. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schwee, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.-W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; Paez-Espino, D.; Jarett, J.; Dunfield, P.; Hedlund, B.P.; Dekas, A.E.; Grasby, S.E.; Brady, A.L.; Dong, H.; Briggs, B.R.; et al. Global metagenomics survey reveals a new bacterial candidate phylum in geothermal springs. Nat. Commun. 2016, 27, 10476. [Google Scholar] [CrossRef] [PubMed]
- Darland, G.; Brock, T.D.; Samsonoff, W.; Conti, S.F. A Thermophilic, Acidophilic Mycoplasma Isolated from a Coal Refuse Pile. Science 1970, 170, 1416–1418. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.J.; Van Houdt, R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, R.; Leys, N.; Vallaeys, T.; Lapidus, A.; et al. The Complete Genome Sequence of Cupriavidus metallidurans Strain CH34, a Master Survivalist in Harsh and Anthropogenic Environments. PLoS ONE 2010, 5, e10433. [Google Scholar] [CrossRef]
- Baker, B.J.; Comolli, L.R.; Dick, G.J.; Hauser, L.J.; Hyatt, D.; Dill, B.D.; Land, M.L.; Verberkmoes, N.C.; Hettich, R.L.; Banfield, J.F. Engimatic, ultrasmall, uncultivated Archaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8806–8811. [Google Scholar] [CrossRef] [PubMed]
- Vorobev, A.V.; DeBoer, W.; Folman, L.B.; Bodelier, P.L.E.; Doronina, N.V.; Suzina, N.E.; Trotsenko, Y.A.; Dedysh, S.N. Methylovirgula ligni gen. nov., sp. nov., an obligately acidophilic, facultatively methylotrophic bacterium with a highly divergent mxaF gene. Int. J. Syst. Evol. Microbiol. 2009, 59, 2538–2545. [Google Scholar] [CrossRef] [PubMed]
- Tarrand, J.J.; Krieg, N.; Dobereiner, J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J. Microbiol. 1978, 24, 967–980. [Google Scholar] [CrossRef]
- Midha, S.; Rigden, D.J.; Siozios, S.; Hurst, G.D.D.; Jackson, A.P. Bodo saltans (Kinetoplastida) is dependent on a novel Paracaedibacter-like endosymbiont that possesses multiple putative toxin-antitoxin systems. ISME J. 2021, 15, 1680–1694. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Tang, G.; Ren, Z.; Zhang, J.; Wang, W.; Qin, X.; Li, K. Ehrlichia, Coxiella and Bartonella infections in rodents from Guizhou Province, Southwest China. Ticks Tick-Borne Dis. 2022, 13, 101974. [Google Scholar] [CrossRef] [PubMed]
- Lehtovirta-Morley, L.E.; Stoecker, K.; Vilcinskas, A.; Prosser, J.I.; Nicol, G.W. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. USA 2011, 108, 15892–15897. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Vejmelkova, D.; Lücker, S.; Streshinskaya, G.M.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Kleerbezem, R.; van Loosdrecht, M.; Muyzer, G.; Daims, H. Nirolancea hollandica gen nov., sp. nov., a chemolithoautotrophic nitrit-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2014, 64, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Suzina, N.E.; Rijpstra, W.I.C.; Damsté, J.S.S.; Dedysh, S.N. Paludibaculum fermentans gen nov., sp. nov., a facultative abaerobe cpable of dissimilatory iron reduction from subdivision 3 of acidobacteria. Int. J. Syst. Evol. Microbiol. 2014, 64, 2857–2864. [Google Scholar] [CrossRef] [PubMed]
- Shelobolina, E.S.; Nevin, K.P.; Blakeney-Hayward, J.D.; Johnsen, C.V.; Plaia, T.W.; Krader, P.; Woodard, T.; Holmes, D.E.; VanPraagh, C.G.; Lovely, D.R. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov. sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 2007, 57, 126–135. [Google Scholar] [CrossRef]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus radiodurans: Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busse, H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov. and Pseudoarthrobacter gen nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar] [PubMed]
- Lin, J.Y.; Russsel, J.A.; Sanders, J.G.; Wertz, J.T. Cephaloticoccus gen. nov., a new genus of ‘Verrucomicrobia’ containing two novel species isolated from Cephalotes ant guts. Int. J. Syst. Evol. Microbiol. 2016, 66, 3034–4040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z.; Patel, B.K.C. Frondicola australicus gen. nov., sp. nov., isolated from decaying leaf litter from a pine forest. Int. J. Syst. Evol. Microbiol. 2007, 57, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T.; Irgens, R.L.; Brenner, D.J.; Pichinoty, F.; De Barjac, H.; Mandel, M.; Asselineau, J. Enhydrobacter aerosaccus gen. nov., sp. nov., a Gas-Vacuolated, Facultatively Anaerobic, Heterotrophic Rod. Int. J. Syst. Bacteriol. 1987, 37, 289–291. [Google Scholar] [CrossRef]
- Manaia, C.; Nogales, B.; Nunes, O. Tepidiphilus margaritifer gen. nov., sp. nov., isolated from a thermophilic aerobic digester. Int. J. Syst. Evol. Microbiol. 2003, 53, 1405–1410. [Google Scholar] [CrossRef]
- Collins, M.D.; Brown, J.; Jones, D. Brachybacterium faecium gen. nov., sp. nov., a Coryneform Bacterium from Poultry Deep Litter. Int. J. Syst. Bacteriol. 1988, 38, 45–48. [Google Scholar] [CrossRef]
- Rossau, R.; Kersters, K.; Falsen, E.; Jantzen, E.; Segers, P.; Union, A.; Nehls, L.; De Ley, J. Oligella, a New Genus Including Oligella urethralis comb. nov. (Formerly Moraxella urethralis) and Oligella ureolytica sp. nov. (Formerly CDC Group IVe): Relationship to Taylorella equigenitalis and Related Taxa. Int. J. Syst. Bacteriol. 1987, 37, 198–210. [Google Scholar] [CrossRef]
- Krumholz, L.R.; Bryant, M.P. Clostridium pfennigii sp. nov. Uses Methoxyl Groups of Monobenzenoids and Produces Butyrate. Int. J. Syst. Bacteriol. 1985, 35, 454–456. [Google Scholar] [CrossRef]
- Kurup, V.R.; Barboriak, J.J.; Fink, J.N.; Lechevalier, M.P. Thermoactinomyces candidus, a new species of thermophilic actinomycetes. Int. J. Syst. Bacteriol. 1975, 25, 150–154. [Google Scholar] [CrossRef]
- Mutschlechner, M.; Lackner, N.; Markt, R.; Salvenmoser, W.; Dunlap, C.A.; Wagner, A.O. Proposal Thermoactinomyces mirandus sp. nov., a filamentous, anaerobic bacterium isolated from a biogas plant. Antonie Van Leeuwenhoek 2021, 114, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Koeck, D.E.; Ludwig, W.; Wanner, G.; Zverlov, V.; Liebl, W.; Schwarz, W. Herbinix hemicellulosilytica gen. nov., sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int. J. Syst. Evol. Microbiol. 2015, 65, 2365–2371. [Google Scholar] [CrossRef]
- Zhang, X.; Tu, B.; Dai, L.-R.; Lawson, P.A.; Zheng, Z.-Z.; Liu, L.-Y.; Deng, Y.; Zhang, H.; Cheng, L. Petroclostridium xylantilyticum gen. nov., sp. nov., a xylan-degrading bacterium isolated from an oilfield, and reclassification of clostridial cluster III members into four novel genera in a new Hungateiclostridiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 3197–3211. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, A.H.; Spring, S.; Schumann, P.; Kroppenstedt, R.M.; Puhakka, J.A. Desulfurispora thermophile gen. nov., sp. nov., a thermophilic, spore-forming sulfate-reducer isolated from a sulfidogenic fluidized-bed reactor. Int. J. Syst. Evol. Microbiol. 2007, 57, 1089–1094. [Google Scholar] [CrossRef]
- Fraj, B.; Ben Hania, W.; Postec, A.; Hamdi, M.; Ollivier, B.; Fardeau, M.-L. Fonticella tunisiensis gen. nov., sp. nov., isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 1947–1950. [Google Scholar] [CrossRef] [PubMed]
- Rheims, H.; Frühling, A.; Schumann, P.; Rohde, M.; Stackebrandt, E. Bacillus silvestris sp. nov., a new member of the genus Bacillus that contains lysine in its cell wall. Int. J. Syst. Bacteriol. 1999, 49, 795–802. [Google Scholar] [CrossRef]
- Krishnamurthi, S.; Chakrabarti, T.; Stackebrandt, E. Re-examination of the taxonomic position of Bacillus silvestris Rheims et al. 1999 and proposal to transfer it to Solibacillus gen. nov. as Solibacillus silvestris comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Zaitsev, G.M.; Tsitko, I.V.; Rainey, F.A.; Trotsenko, Y.A.; Uotila, J.S.; Stackebrandt, E.; Salkinoja-Salonen, M.S. New aerobic ammonium-dependent obligately oxalotrophic bacteria: Description of Ammoniphilus oxalaticus gen. nov., sp. nov. and Ammoniphilus oxalivorans gen. nov., sp. nov. Int. J. Syst. Bacteriol. 1998, 48, 151–163. [Google Scholar] [CrossRef] [PubMed]
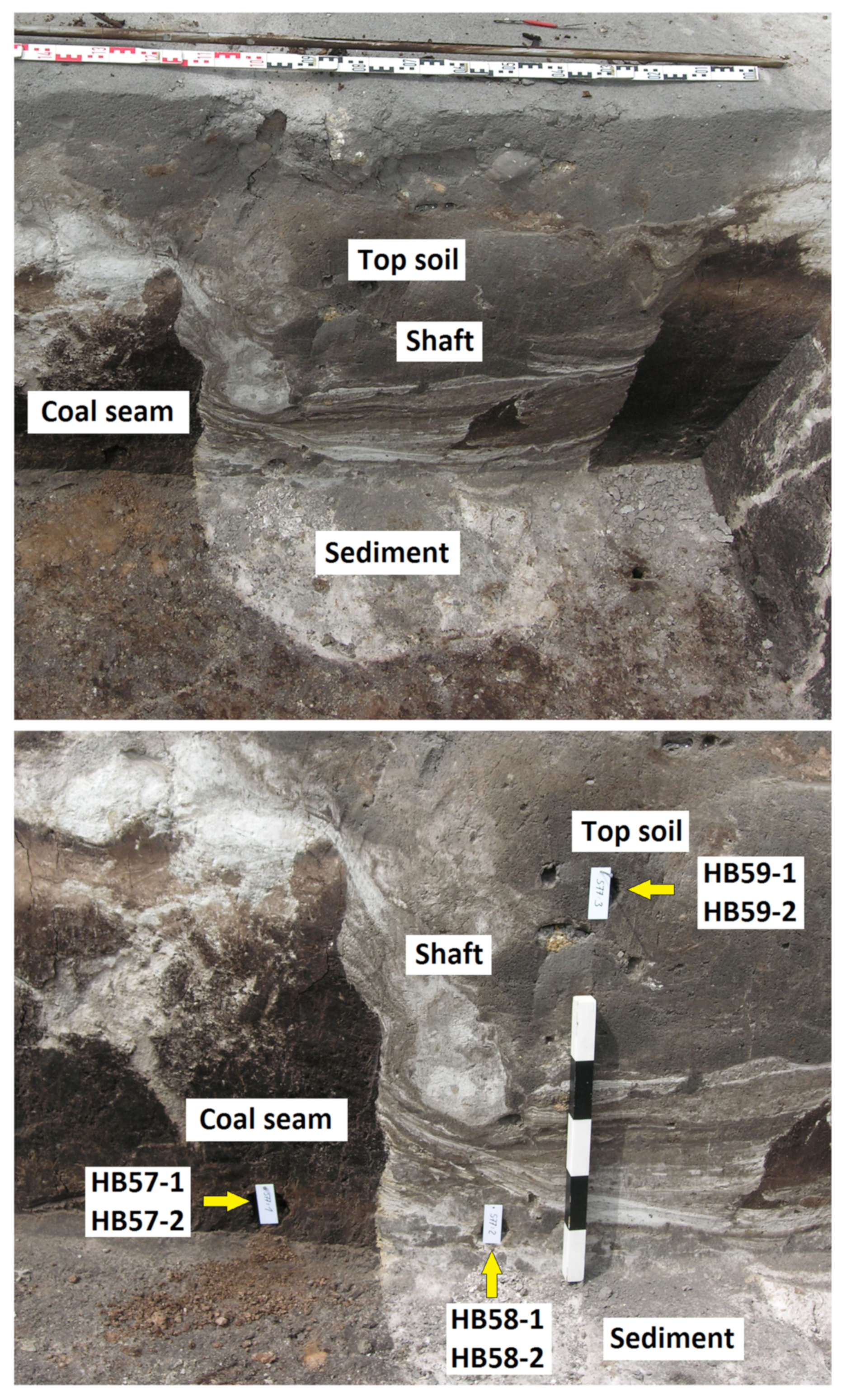





| Depth | pH | Conductivity | ||
|---|---|---|---|---|
| HB57-1, HB57-2 | coal seam | 1.6 m | 4.22 | 114.8 (µS/cm) |
| HB58-1, HB58-2 | bright sediment | 1.6 m | 4.09 | 43.3 (µS/cm) |
| HB59-1, HB59-2 | topsoil sediment | 1.0 m | 4.05 | 45.2 (µS/cm) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehrhardt, L.; Günther, P.M.; Böhme, M.; Köhler, J.M.; Cao, J. Three Soil Bacterial Communities from an Archaeological Excavation Site of an Ancient Coal Mine near Bennstedt (Germany) Characterized by 16S r-RNA Sequencing. Environments 2022, 9, 115. https://doi.org/10.3390/environments9090115
Ehrhardt L, Günther PM, Böhme M, Köhler JM, Cao J. Three Soil Bacterial Communities from an Archaeological Excavation Site of an Ancient Coal Mine near Bennstedt (Germany) Characterized by 16S r-RNA Sequencing. Environments. 2022; 9(9):115. https://doi.org/10.3390/environments9090115
Chicago/Turabian StyleEhrhardt, Linda, P. Mike Günther, Manfred Böhme, J. Michael Köhler, and Jialan Cao. 2022. "Three Soil Bacterial Communities from an Archaeological Excavation Site of an Ancient Coal Mine near Bennstedt (Germany) Characterized by 16S r-RNA Sequencing" Environments 9, no. 9: 115. https://doi.org/10.3390/environments9090115






