A State-of-the-Art Review on SARS-CoV-2 Virus Removal Using Different Wastewater Treatment Strategies
Abstract
:1. Introduction
2. Review Methodology
3. Detection Protocol of SARS-CoV-2 RNA in Water Matrices
4. Status of SARS-CoV-2 RNA in Sewage
4.1. Comparison of Different Titers of SARS-CoV-2 RNA in Sewage
4.2. Possible Fate of COVID-19 RNA after Ingestion
5. Wastewater Treatment Methods to Eliminate the SARS-CoV-2 RNA
5.1. Need of Wastewater Treatment
5.2. Membrane-Based Technologies
5.2.1. Polymeric Membrane
5.2.2. Ceramic Membrane
5.2.3. Membrane Bioreactor (MBR)
5.3. Disinfection-Based Strategies
5.3.1. Chlorine-Based Disinfectants
5.3.2. Ultraviolet Radiation/Solar-Assisted Inactivation
5.3.3. Ozonation
5.3.4. Hydrogen Peroxide
5.3.5. Nanomaterials and Photocatalysis
5.3.6. Pond/Algal Systems
5.3.7. Advanced Oxidation Processes (AOPs)
6. Conclusions and Recommendations
- To establish a standard method for reducing the volume to obtain the highest possible amount of RNA.
- The regular chlorination of wastewater treatment plants can inactivate a broad range of viruses and SARS-CoV-2, but the legal dose should be considered because of its side effects.
- To evaluate the degree of contamination in raw agricultural food products when reusing water for irrigation.
- Hospitals should immediately adopt advanced progressive technologies to manage the epidemiology of SARS-CoV-2 through quick approval. Wastewater surveillance of disease-causing agents in hospitals, thus, is to be urgently established, where the disinfectant rule could be easily implemented.
- In low-sanitation nations, decentralized wastewater treatment systems should be improved. Furthermore, chlorination, before the wastewater is discharged into rivers and thus, into the ocean, is the easiest possible method.
- Future work must focus on implementing the selected actions for the treatment of the wastewater released from the COVID-19 hospitals and self-quarantine centers to better regulate future waves of SARS-CoV-2.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef] [PubMed]
- Chik, A.H.; Glier, M.B.; Servos, M.; Mangat, C.S.; Pang, X.-L.; Qiu, Y.; D’Aoust, P.M.; Burnet, J.-B.; Delatolla, R.; Dorner, S. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada. J. Environ. Sci. 2021, 107, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Mousazadeh, M.; Ashoori, R.; Paital, B.; Kabdaşlı, I.; Frontistis, Z.; Hashemi, M.; Sandoval, M.A.; Sherchan, S.; Das, K.; Emamjomeh, M.M. Wastewater Based Epidemiology Perspective as a Faster Protocol for Detecting Coronavirus RNA in Human Populations: A Review with Specific Reference to SARS-CoV-2 Virus. Pathogens 2021, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Li, X.; Sunita, K.; Lokhandwala, S.; Gautam, P.; Suresh, S.; Sarma, H.; Vellingiri, B.; Dey, A.; Bontempi, E. SARS-CoV-2 and other pathogens in municipal wastewater, landfill leachate, and solid waste: A review about virus surveillance, infectivity, and inactivation. Environ. Res. 2021, 203, 111839. [Google Scholar] [CrossRef]
- Balboa, S.; Mauricio-Iglesias, M.; Rodriguez, S.; Martínez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-CoV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021, 772, 145268. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2021, 40, 101815. [Google Scholar] [CrossRef]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar]
- Sangkham, S. A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral particles. J. Environ. Manag. 2021, 299, 113563. [Google Scholar] [CrossRef]
- Serra-Compte, A.; González, S.; Arnaldos, M.; Berlendis, S.; Courtois, S.; Loret, J.F.; Schlosser, O.; Yáñez, A.M.; Soria-Soria, E.; Fittipaldi, M. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021, 202, 117435. [Google Scholar] [CrossRef] [PubMed]
- Parsa, S.M.; Momeni, S.; Hemmat, A.; Afrand, M. Effectiveness of solar water disinfection in the era of COVID-19 (SARS-CoV-2) pandemic for contaminated water/wastewater treatment considering UV effect and temperature. J. Water Process Eng. 2021, 43, 102224. [Google Scholar] [CrossRef]
- Xagoraraki, I.; O’Brien, E. Wastewater-based epidemiology for early detection of viral outbreaks. In Women in Water Quality; Tietjen, J.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 75–97. [Google Scholar]
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef]
- Sinclair, R.G.; Choi, C.Y.; Riley, M.R.; Gerba, C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008, 65, 249. [Google Scholar] [PubMed]
- Hellmér, M.; Paxéus, N.; Magnius, L.; Enache, L.; Arnholm, B.; Johansson, A.; Bergström, T.; Norder, H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014, 80, 6771–6781. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Minard-Smith, A.; Catlin, L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total Environ. 2021, 789, 147829. [Google Scholar] [CrossRef] [PubMed]
- Giacobbo, A.; Rodrigues, M.A.S.; Ferreira, J.Z.; Bernardes, A.M.; de Pinho, M.N. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total Environ. 2021, 774, 145721. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Guo, T.; Zhen, B.; Kong, Q.; Yi, B.; Li, Z.; Song, N.; Jin, M.; Xiao, W. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci. Technol. 2005, 52, 213–221. [Google Scholar] [CrossRef]
- Betancourt, W.Q.; Schmitz, B.W.; Innes, G.K.; Prasek, S.M.; Brown, K.M.P.; Stark, E.R.; Foster, A.R.; Sprissler, R.S.; Harris, D.T.; Sherchan, S.P. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408. [Google Scholar] [CrossRef]
- Miyani, B.; Fonoll, X.; Norton, J.; Mehrotra, A.; Xagoraraki, I. SARS-CoV-2 in Detroit wastewater. J. Environ. Eng. 2020, 146, 06020004. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems 2020, 5, e00614–e00620. [Google Scholar] [CrossRef]
- Nasseri, S.; Yavarian, J.; Baghani, A.N.; Azad, T.M.; Nejati, A.; Nabizadeh, R.; Hadi, M.; Jandaghi, N.Z.S.; Vakili, B.; Vaghefi, S.K.A. The presence of SARS-CoV-2 in raw and treated wastewater in 3 cities of Iran: Tehran, Qom and Anzali during coronavirus disease 2019 (COVID-19) outbreak. J. Environ. Health Sci. Eng. 2021, 19, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Orschler, L.; Lackner, S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021, 11, 5372. [Google Scholar] [CrossRef] [PubMed]
- Saththasivam, J.; El-Malah, S.S.; Gomez, T.A.; Jabbar, K.A.; Remanan, R.; Krishnankutty, A.K.; Ogunbiyi, O.; Rasool, K.; Ashhab, S.; Rashkeev, S. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci. Total Environ. 2021, 774, 145608. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Marechal, V.; Mouchel, J.-M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv 2020. medRxiv:2020.04.12.20062679. [Google Scholar]
- Zhang, D.; Ling, H.; Huang, X.; Li, J.; Li, W.; Yi, C.; Zhang, T.; Jiang, Y.; He, Y.; Deng, S. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020, 741, 140445. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.G.; Gebhard, L.G.; Carballeda, J.M.; Aiello, I.; Recalde, E.; Terny, G.; Ambrosolio, S.; L’Arco, G.; Konfino, J.; Brardinelli, J.I. SARS-CoV-2 surveillance in untreated wastewater: First detection in a low-resource community in Buenos Aires, Argentina. medRxiv 2020. medRxiv:2020.10.21.20215434. [Google Scholar]
- Sharif, S.; Ikram, A.; Khurshid, A.; Salman, M.; Mehmood, N.; Arshad, Y.; Ahmad, J.; Safdar, R.M.; Angez, M.; Alam, M.M. Detection of SARs-CoV-2 in wastewater, using the existing environmental surveillance network: An epidemiological gateway to an early warning for COVID-19 in communities. medRxiv 2020. medRxiv:2020.06.03.20121426. [Google Scholar]
- Spurbeck, R.R.; Minard-Smith, A.T.; Catlin, L.A. Applicability of Neighborhood and Building Scale Wastewater-Based Genomic Epidemiology to Track the SARS-CoV-2 Pandemic and other Pathogens. medRxiv 2021. medRxiv:2021.02.18.21251939. [Google Scholar]
- Gonçalves, J.; Koritnik, T.; Mioč, V.; Trkov, M.; Bolješič, M.; Berginc, N.; Prosenc, K.; Kotar, T.; Paragi, M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021, 755, 143226. [Google Scholar] [CrossRef]
- Kocamemi, B.A.; Kurt, H.; Hacıoglu, S.; Yaralı, C.; Saatci, A.M.; Pakdemirli, B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. medRxiv 2020. medRxiv:2020.05.03.20089417. [Google Scholar]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Bar-Or, I.; Yaniv, K.; Shagan, M.; Ozer, E.; Weil, M.; Indenbaum, V.; Elul, M.; Erster, O.; Mendelson, E.; Mannasse, B. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: A proof-of-concept for quantitative environmental surveillance. Front. Public Health 2021, 9, 561710. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef]
- Trottier, J.; Darques, R.; Mouheb, N.A.; Partiot, E.; Bakhache, W.; Deffieu, M.S.; Gaudin, R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health 2020, 10, 100157. [Google Scholar] [CrossRef]
- Mlejnkova, H.; Sovova, K.; Vasickova, P.; Ocenaskova, V.; Jasikova, L.; Juranova, E. Preliminary study of SARS-CoV-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health 2020, 17, 5508. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Baldovin, T.; Amoruso, I.; Fonzo, M.; Buja, A.; Baldo, V.; Cocchio, S.; Bertoncello, C. SARS-CoV-2 RNA detection and persistence in wastewater samples: An experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy). Sci. Total Environ. 2021, 760, 143329. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.-J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef]
- Kolarević, S.; Micsinai, A.; Szántó-Egész, R.; Lukács, A.; Kračun-Kolarević, M.; Lundy, L.; Kirschner, A.K.; Farnleitner, A.H.; Djukic, A.; Čolić, J. Detection of SARS-CoV-2 RNA in the Danube River in Serbia associated with the discharge of untreated wastewaters. Sci. Total Environ. 2021, 783, 146967. [Google Scholar] [CrossRef]
- Hata, A.; Hara-Yamamura, H.; Meuchi, Y.; Imai, S.; Honda, R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021, 758, 143578. [Google Scholar] [CrossRef]
- Kitamura, K.; Sadamasu, K.; Muramatsu, M.; Yoshida, H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021, 763, 144587. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schörner, M.A.; Barazzetti, F.H.; Christoff, A.P.; de Oliveira, L.F.V. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021, 778, 146198. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef]
- Randazzo, W.; Cuevas-Ferrando, E.; Sanjuán, R.; Domingo-Calap, P.; Sánchez, G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health 2020, 230, 113621. [Google Scholar] [CrossRef] [PubMed]
- Prado, T.; Fumian, T.M.; Mannarino, C.F.; Resende, P.C.; Motta, F.C.; Eppinghaus, A.L.F.; do Vale, V.H.C.; Braz, R.M.S.; de Andrade, J.d.S.R.; Maranhão, A.G. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021, 191, 116810. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021, 750, 141711. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.W.; Ibrahim, Y.; Daou, M.; Kannout, H.; Jan, N.; Lopes, A.; Alsafar, H.; Yousef, A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021, 764, 142929. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef]
- Castiglioni, S.; Thomas, K.V.; Kasprzyk-Hordern, B.; Vandam, L.; Griffiths, P. Testing wastewater to detect illicit drugs: State of the art, potential and research needs. Sci. Total Environ. 2014, 487, 613–620. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Alygizakis, N.; Markou, A.N.; Rousis, N.I.; Galani, A.; Avgeris, M.; Adamopoulos, P.G.; Scorilas, A.; Lianidou, E.S.; Paraskevis, D.; Tsiodras, S. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: Protocols and future perspectives. TrAC Trends Anal. Chem. 2020, 134, 116125. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020, 8, 104306. [Google Scholar] [CrossRef] [PubMed]
- Beattie, R.E.; Blackwood, A.D.; Clerkin, T.; Dinga, C.; Noble, R.T. Evaluating the impact of sample storage, handling, and technical ability on the decay and recovery of SARS-CoV-2 in wastewater. PLoS ONE 2022, 17, e0270659. [Google Scholar] [CrossRef]
- Bivins, A.; North, D.; Wu, Z.; Shaffer, M.; Ahmed, W.; Bibby, K. Within-and between-day variability of SARS-CoV-2 RNA in municipal wastewater during periods of varying COVID-19 prevalence and positivity. ACS ES&T Water 2021, 1, 2097–2108. [Google Scholar]
- Ahmed, W.; Bivins, A.; Simpson, S.L.; Smith, W.J.; Metcalfe, S.; McMinn, B.; Symonds, E.M.; Korajkic, A. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Sci. Total Environ. 2021, 799, 149386. [Google Scholar] [CrossRef] [PubMed]
- Sapula, S.A.; Whittall, J.J.; Pandopulos, A.J.; Gerber, C.; Venter, H. An optimized and robust PEG precipitation method for detection of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 785, 147270. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Deng, Y.; Xu, X.; Li, S.; Zhang, Y.; Ding, J.; On, H.Y.; Lai, J.C.; Yau, C.I.; Chin, A.W. Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci. Total Environ. 2022, 824, 153687. [Google Scholar] [CrossRef]
- Pérez-Cataluña, A.; Cuevas-Ferrando, E.; Randazzo, W.; Falcó, I.; Allende, A.; Sánchez, G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 758, 143870. [Google Scholar] [CrossRef]
- Hasing, M.; Yu, J.; Qiu, Y.; Maal-Bared, R.; Bhavanam, S.; Lee, B.; Hrudey, S.; Pang, X. Comparison of detecting and quantitating SARS-CoV-2 in wastewater using moderate-speed centrifuged solids versus an ultrafiltration method. Water 2021, 13, 2166. [Google Scholar] [CrossRef]
- Philo, S.E.; Keim, E.K.; Swanstrom, R.; Ong, A.Q.; Burnor, E.A.; Kossik, A.L.; Harrison, J.C.; Demeke, B.A.; Zhou, N.A.; Beck, N.K. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021, 760, 144215. [Google Scholar] [CrossRef]
- Cervantes-Avilés, P.; Moreno-Andrade, I.; Carrillo-Reyes, J. Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19. J. Water Process Eng. 2021, 40, 101947. [Google Scholar] [CrossRef]
- Wozniak, A.; Cerda, A.; Ibarra-Henriquez, C.; Sebastian, V.; Armijo, G.; Lamig, L.; Miranda, C.; Lagos, M.; Solari, S.; Guzmán, A.M. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci. Rep. 2020, 10, 16608. [Google Scholar] [CrossRef]
- Tavares, L.; Alves, P.M.; Ferreira, R.B.; Santos, C.N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res. Notes 2011, 4, 3. [Google Scholar]
- Barril, P.A.; Pianciola, L.A.; Mazzeo, M.; Ousset, M.J.; Jaureguiberry, M.V.; Alessandrello, M.; Sánchez, G.; Oteiza, J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2021, 756, 144105. [Google Scholar] [CrossRef] [PubMed]
- Canh, V.D.; Torii, S.; Yasui, M.; Kyuwa, S.; Katayama, H. Capsid integrity RT-qPCR for the selective detection of intact SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 791, 148342. [Google Scholar] [CrossRef] [PubMed]
- Masindi, V.; Foteinis, S.; Nduli, K.; Akinwekomi, V. Systematic assessment of SARS-CoV-2 virus in wastewater, rivers and drinking water—A catchment-wide appraisal. Sci. Total Environ. 2021, 800, 149298. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Gómez, L.; Sanseverino, I.; Niegowska, M.; Roka, E.; Pedraccini, R.; Vargha, M.; Lettieri, T. SARS-CoV-2 detection in wastewater using multiplex quantitative PCR. Sci. Total Environ. 2021, 797, 148890. [Google Scholar] [CrossRef]
- Palmer, E.J.; Maestre, J.P.; Jarma, D.; Lu, A.; Willmann, E.; Kinney, K.A.; Kirisits, M.J. Development of a reproducible method for monitoring SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 799, 149405. [Google Scholar] [CrossRef]
- Tomasino, M.P.; Semedo, M.; Vieira, P.; Ferraz, E.; Rocha, A.; Carvalho, M.F.; Magalhães, C.; Mucha, A.P. SARS-CoV-2 RNA detected in urban wastewater from Porto, Portugal: Method optimization and continuous 25-week monitoring. Sci. Total Environ. 2021, 792, 148467. [Google Scholar] [CrossRef]
- Wurtzer, S.; Waldman, P.; Ferrier-Rembert, A.; Frenois-Veyrat, G.; Mouchel, J.-M.; Boni, M.; Maday, Y.; Marechal, V.; Moulin, L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 2021, 198, 117183. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef]
- Kumar, N.; Shetti, N.P.; Jagannath, S.; Aminabhavi, T.M. Electrochemical sensors for the detection of SARS-CoV-2 virus. Chem. Eng. J. 2022, 430, 132966. [Google Scholar] [CrossRef]
- Dignan, L.M.; Turiello, R.; Layne, T.R.; O’Connell, K.C.; Hickey, J.; Chapman, J.; Poulter, M.D.; Landers, J.P. An ultrafast SARS-CoV-2 virus enrichment and extraction method compatible with multiple modalities for RNA detection. Anal. Chim. Acta 2021, 1180, 338846. [Google Scholar] [CrossRef]
- Lu, D.; Zhu, D.Z.; Gan, H.; Yao, Z.; Fu, Q.; Zhang, X.J. Prospects and challenges of using electrochemical immunosensors as an alternative detection method for SARS-CoV-2 wastewater-based epidemiology. Sci. Total Environ. 2021, 777, 146239. [Google Scholar] [CrossRef]
- Peng, Y.; Pan, Y.; Sun, Z.; Li, J.; Yi, Y.; Yang, J.; Li, G. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. 2021, 186, 113309. [Google Scholar] [CrossRef] [PubMed]
- Pierce-Ruiz, C.; Santana, W.I.; Sutton, W.J.; Fischler, D.A.; Cooper, H.C.; Marc, L.R.; Barr, J.R.; Williams, T.L. Quantification of SARS-CoV-2 spike and nucleocapsid proteins using isotope dilution tandem mass spectrometry. Vaccine 2021, 39, 5106–5115. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, H.; Yang, Z. Can a Paper-Based Device Trace COVID-19 Sources with Wastewater-Based Epidemiology? ACS Publications: Washington, DC, USA, 2020. [Google Scholar]
- Lee, W.L.; Imakaev, M.; Armas, F.; McElroy, K.A.; Gu, X.; Duvallet, C.; Chandra, F.; Chen, H.; Leifels, M.; Mendola, S. Quantitative SARS-CoV-2 Alpha Variant B. 1.1. 7 Tracking in Wastewater by Allele-Specific RT-qPCR. Environ. Sci. Technol. Lett. 2021, 8, 675–682. [Google Scholar] [CrossRef]
- Tiwari, A.; Phan, N.; Tandukar, S.; Ashoori, R.; Thakali, O.; Mousazadesh, M.; Dehghani, M.H.; Sherchan, S.P. Persistence and occurrence of SARS-CoV-2 in water and wastewater environments: A review of the current literature. Environ. Sci. Pollut. Res. 2021, 1–11. [Google Scholar] [CrossRef]
- Polo, D.; Lois, M.; Fernández-Núñez, M.T.; Romalde, J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021, 786, 147534. [Google Scholar] [CrossRef]
- Topare, N.S.; Attar, S.; Manfe, M.M. Sewage/wastewater treatment technologies: A review. Sci. Revs. Chem. Commun 2011, 1, 18–24. [Google Scholar]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Curtis, K.; Keeling, D.; Yetka, K.; Larson, A.; Gonzalez, R. Wastewater SARS-CoV-2 concentration and loading variability from grab and 24-hour composite samples. medRxiv 2020. medRxiv:2020.07.10.20150607. [Google Scholar]
- Kumar, M.; Kuroda, K.; Patel, A.K.; Patel, N.; Bhattacharya, P.; Joshi, M.; Joshi, C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021, 754, 142329. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Núñez-Acuña, G.; Valenzuela-Miranda, D.; Benaventel, B.P.; Sáez-Vera, C.; Urrutia, H.; Novoa, B.; Figueras, A.; Roberts, S. The wastewater microbiome: A novel insight for COVID-19 surveillance. Sci. Total Environ. 2021, 764, 142867. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; North, D.; Ahmad, A.; Ahmed, W.; Alm, E.; Been, F.; Bhattacharya, P.; Bijlsma, L.; Boehm, A.B.; Brown, J. Wastewater-Based Epidemiology: GLOBAL Collaborative to Maximize Contributions in the Fight against COVID-19; ACS Publications: Washington, DC, USA, 2020. [Google Scholar]
- Wang, J.; Feng, H.; Zhang, S.; Ni, Z.; Ni, L.; Chen, Y.; Zhuo, L.; Zhong, Z.; Qu, T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020, 94, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Natarajan, A.; Han, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Wolfe, M.; Singh, U.; Jagannathan, P.; Pinsky, B.A.; Boehm, A. Standardized preservation, extraction and quantification techniques for detection of fecal SARS-CoV-2 RNA. Nat. Commun. 2021, 12, 5753. [Google Scholar] [CrossRef] [PubMed]
- Saawarn, B.; Hait, S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J. Environ. Chem. Eng. 2021, 9, 104870. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, L.; Deng, Q.; Zhang, G.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020, 92, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Quilliam, R.S.; Weidmann, M.; Moresco, V.; Purshouse, H.; O’Hara, Z.; Oliver, D.M. COVID-19: The environmental implications of shedding SARS-CoV-2 in human faeces. Environ. Int. 2020, 140, 105790. [Google Scholar] [CrossRef]
- Zhang, N.; Gong, Y.; Meng, F.; Shi, Y.; Wang, J.; Mao, P.; Chuai, X.; Bi, Y.; Yang, P.; Wang, F. Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci. China Life Sci. 2021, 64, 486–488. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Xu, K.; Chen, Y.; Yuan, J.; Yi, P.; Ding, C.; Wu, W.; Li, Y.; Ni, Q.; Zou, R.; Li, X.; et al. Factors Associated With Prolonged Viral RNA Shedding in Patients with Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2020, 71, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.-H.; Ni, W.; Wu, Q.; Li, W.-J.; Li, G.-J.; Wang, W.-D.; Tong, J.-N.; Song, X.-F.; Wing-Kin Wong, G.; Xing, Q.-S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020, 53, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Róka, E.; Khayer, B.; Kis, Z.; Kovács, L.B.; Schuler, E.; Magyar, N.; Málnási, T.; Oravecz, O.; Pályi, B.; Pándics, T. Ahead of the second wave: Early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 2021, 786, 147398. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.K.; Sathyamurthy, R.; Velraj, R.; Lynch, I.; Saidur, R.; Pandey, A.; Sharshir, S.W.; Hwang, J.-Y.; GaneshKumar, P. Secondary transmission of SARS-CoV-2 through wastewater: Concerns and tactics for treatment to effectively control the pandemic. J. Environ. Manag. 2021, 290, 112668. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, C.A.R.; Hilares, R.T.; Andrade, G.d.J.C.; Mogrovejo-Valdivia, A.; Tanaka, D.A.P. Emerging contaminants, SARS-COV-2 and wastewater treatment plants, new challenges to confront: A short review. Bioresour. Technol. Rep. 2021, 15, 100731. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Street, R.; Malema, S.; Mahlangeni, N.; Mathee, A. COVID-19 wastewater surveillance: An African perspective. Sci. Total Environ. 2020, 743, 140719. [Google Scholar] [CrossRef]
- Tortajada, C.; Biswas, A.K. COVID-19 heightens water problems around the world. Water Int. 2020, 45, 441–442. [Google Scholar] [CrossRef]
- Mohan, S.V.; Hemalatha, M.; Kopperi, H.; Ranjith, I.; Kumar, A.K. SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021, 405, 126893. [Google Scholar] [CrossRef]
- Paul, D.; Kolar, P.; Hall, S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. NPJ Clean Water 2021, 4, 7. [Google Scholar] [CrossRef]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.-S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.; Sarmah, A.K.; Chao, H.-P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Li, J.-S.; Jin, M.; Zhen, B.; Kong, Q.-X.; Song, N.; Xiao, W.-J.; Yin, J.; Wei, W.; Wang, G.-J. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef]
- de Oliveira, L.C.; Torres-Franco, A.F.; Lopes, B.C.; da Silva Santos, B.S.Á.; Costa, E.A.; Costa, M.S.; Reis, M.T.P.; Melo, M.C.; Polizzi, R.B.; Teixeira, M.M. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021, 195, 117002. [Google Scholar] [CrossRef] [PubMed]
- Collivignarelli, M.C.; Collivignarelli, C.; Miino, M.C.; Abbà, A.; Pedrazzani, R.; Bertanza, G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020, 143, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Lesimple, A.; Jasim, S.Y.; Johnson, D.J.; Hilal, N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. J. Water Process Eng. 2020, 38, 101544. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- Abu Ali, H.; Yaniv, K.; Bar-Zeev, E.; Chaudhury, S.; Shagan, M.; Lakkakula, S.; Ronen, Z.; Kushmaro, A.; Nir, O. Tracking SARS-CoV-2 RNA through the wastewater treatment process. ACS ES&T Water 2021, 1, 1161–1167. [Google Scholar]
- Eloffy, M.; El-Sherif, D.M.; Abouzid, M.; Abd Elkodous, M.; El-nakhas, H.S.; Sadek, R.F.; Ghorab, M.A.; Al-Anazi, A.; El-Sayyad, G.S. Proposed approaches for coronaviruses elimination from wastewater: Membrane techniques and nanotechnology solutions. Nanotechnol. Rev. 2022, 11, 1–25. [Google Scholar] [CrossRef]
- Viralzone. 2021. Available online: https://viralzone.expasy.org/5216 (accessed on 3 August 2021).
- Pendergast, M.M.; Hoek, E.M. A review of water treatment membrane nanotechnologies. Energy Environ. Sci. 2011, 4, 1946–1971. [Google Scholar] [CrossRef]
- Im, D.; Nakada, N.; Kato, Y.; Aoki, M.; Tanaka, H. Pretreatment of ceramic membrane microfiltration in wastewater reuse: A comparison between ozonation and coagulation. J. Environ. Manag. 2019, 251, 109555. [Google Scholar] [CrossRef]
- Bartels, J.; Batista, A.G.; Kroll, S.; Maas, M.; Rezwan, K. Hydrophobic ceramic capillary membranes for versatile virus filtration. J. Membr. Sci. 2019, 570, 85–92. [Google Scholar] [CrossRef]
- Zielińska, M.; Galik, M. Use of Ceramic Membranes in a Membrane Filtration Supported by Coagulation for the Treatment of Dairy Wastewater. Water Air Soil Pollut. 2017, 228, 173. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hayward, K.; Khan, S.J.; Örmeci, B.; Pillay, S.; Rose, J.B.; Thanikal, J.V.; Zhang, T. Role of wastewater treatment in COVID-19 control. Water Qual. Res. J. 2021, 56, 68–82. [Google Scholar] [CrossRef]
- Majumder, A.; Gupta, A.K.; Ghosal, P.S.; Varma, M. A review on hospital wastewater treatment: A special emphasis on occurrence and removal of pharmaceutically active compounds, resistant microorganisms, and SARS-CoV-2. J. Environ. Chem. Eng. 2020, 9, 104812. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, J.; Ye, D.; Yan, X.; Zhang, Y.; Yang, W.; Li, X.; Wang, J.; Zhang, L.; Pan, L. Disinfection technology of hospital wastes and wastewater: Suggestions for disinfection strategy during coronavirus Disease 2019 (COVID-19) pandemic in China. Environ. Pollut. 2020, 262, 114665. [Google Scholar] [CrossRef]
- Lee, H.-W.; Lee, H.-M.; Yoon, S.-R.; Kim, S.H.; Ha, J.-H. Pretreatment with propidium monoazide/sodium lauroyl sarcosinate improves discrimination of infectious waterborne virus by RT-qPCR combined with magnetic separation. Environ. Pollut. 2018, 233, 306–314. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Kapoor, D.; Dhanjal, D.S.; Bhatia, D.; Jan, S.; Singh, N.; Romero, R.; Ramamurthy, P.C.; Singh, J. Detection and disinfection of COVID-19 virus in wastewater. Environ. Chem. Lett. 2021, 19, 1917–1933. [Google Scholar] [CrossRef]
- Alahdal, H.M.; AlYahya, S.; Ameen, F.; Sonbol, H.; Alomary, M.N. A review on Saudi Arabian wastewater treatment facilities and available disinfection methods: Implications to SARS-CoV-2 control. J. King Saud Univ. Sci. 2021, 33, 101574. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, Q.; Lee, B.E.; Ruecker, N.J.; Neumann, N.F.; Ashbolt, N.J.; Pang, X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018, 147, 73–81. [Google Scholar] [CrossRef]
- Cheng, S.; Ge, Y.; Lee, Y.; Yang, X. Prediction of Photolysis Kinetics of Viral Genomes under UV254 Irradiation to Estimate Virus Infectivity Loss. Water Res. 2021, 198, 117165. [Google Scholar] [CrossRef]
- Nelson, K.L.; Boehm, A.B.; Davies-Colley, R.J.; Dodd, M.C.; Kohn, T.; Linden, K.G.; Liu, Y.; Maraccini, P.A.; McNeill, K.; Mitch, W.A. Sunlight-mediated inactivation of health-relevant microorganisms in water: A review of mechanisms and modeling approaches. Environ. Sci. Process. Impacts 2018, 20, 1089–1122. [Google Scholar] [CrossRef]
- Tizaoui, C. Ozone: A potential oxidant for COVID-19 virus (SARS-CoV-2). Ozone Sci. Eng. 2020, 42, 378–385. [Google Scholar] [CrossRef]
- Wolf, C.; von Gunten, U.; Kohn, T. Kinetics of inactivation of waterborne enteric viruses by ozone. Environ. Sci. Technol. 2018, 52, 2170–2177. [Google Scholar] [CrossRef] [PubMed]
- Paraskeva, P.; Graham, N.J. Ozonation of municipal wastewater effluents. Water Environ. Res. 2002, 74, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Volkoff, S.J.; Carlson, T.J.; Leik, K.; Smith, J.J.; Graves, D.; Dennis, P.; Aris, T.; Cuthbertson, D.; Holmes, A.; Craig, K. Demonstrated SARS-CoV-2 Surface Disinfection Using Ozone. Ozone Sci. Eng. 2021, 43, 296–305. [Google Scholar] [CrossRef]
- Kataki, S.; Chatterjee, S.; Vairale, M.G.; Sharma, S.; Dwivedi, S.K. Concerns and strategies for wastewater treatment during COVID-19 pandemic to stop plausible transmission. Resour. Conserv. Recycl. 2020, 164, 105156. [Google Scholar] [CrossRef]
- McDonnell, G. The use of hydrogen peroxide for disinfection and sterilization applications. PATAI’S Chem. Funct. Groups 2009, 1–34. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Q.; Chen, L.; Wang, J.; Cheng, R. Photocatalytic membrane reactor (PMR) for virus removal in water: Performance and mechanisms. Chem. Eng. J. 2015, 277, 124–129. [Google Scholar] [CrossRef]
- Rahaman, M.S.; Vecitis, C.D.; Elimelech, M. Electrochemical carbon-nanotube filter performance toward virus removal and inactivation in the presence of natural organic matter. Environ. Sci. Technol. 2012, 46, 1556–1564. [Google Scholar] [CrossRef]
- Han, W.; Zhang, P.H.; Cao, W.C.; Yang, D.L.; Taira, S.; Okamoto, Y.; Arai, J.I.; Yan, X.Y. The inactivation effect of photocatalytic titanium apatite filter on SARS virus. Prog. Biochem. Biophys. 2004, 31, 982–985. [Google Scholar]
- Verbyla, M.E.; Mihelcic, J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015, 71, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Delanka-Pedige, H.M.; Cheng, X.; Munasinghe-Arachchige, S.P.; Abeysiriwardana-Arachchige, I.S.; Xu, J.; Nirmalakhandan, N.; Zhang, Y. Metagenomic insights into virus removal performance of an algal-based wastewater treatment system utilizing Galdieria sulphuraria. Algal Res. 2020, 47, 101865. [Google Scholar] [CrossRef]
- Saba, B.; Hasan, S.W.; Kjellerup, B.V.; Christy, A.D. Capacity of existing wastewater treatment plants to treat SARS-CoV-2. A review. Bioresour. Technol. Rep. 2021, 15, 100737. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Tang, W.; Yu, L.; Liu, Z.; Liu, Y.; Xia, H.; Zhang, H.; Chen, S.; Wu, J.; Cui, X. Inactivating SARS-CoV-2 by electrochemical oxidation. Sci. Bull. 2021, 66, 720–726. [Google Scholar] [CrossRef]
- Kokkinos, P.; Venieri, D.; Mantzavinos, D. Advanced Oxidation Processes for Water and Wastewater Viral Disinfection. A Systematic Review. Food Environ. Virol. 2021, 13, 283–302. [Google Scholar] [CrossRef] [PubMed]
- Škulcová, A.B.; Tamášová, K.; Staňová, A.V.; Bírošová, L.; Krahulcová, M.; Gál, M.; Konečná, B.; Janíková, M.; Celec, P.; Grabicová, K. Effervescent ferrate (VI)-based tablets as an effective means for removal SARS-CoV-2 RNA, pharmaceuticals and resistant bacteria from wastewater. J. Water Process Eng. 2021, 43, 102223. [Google Scholar] [CrossRef]
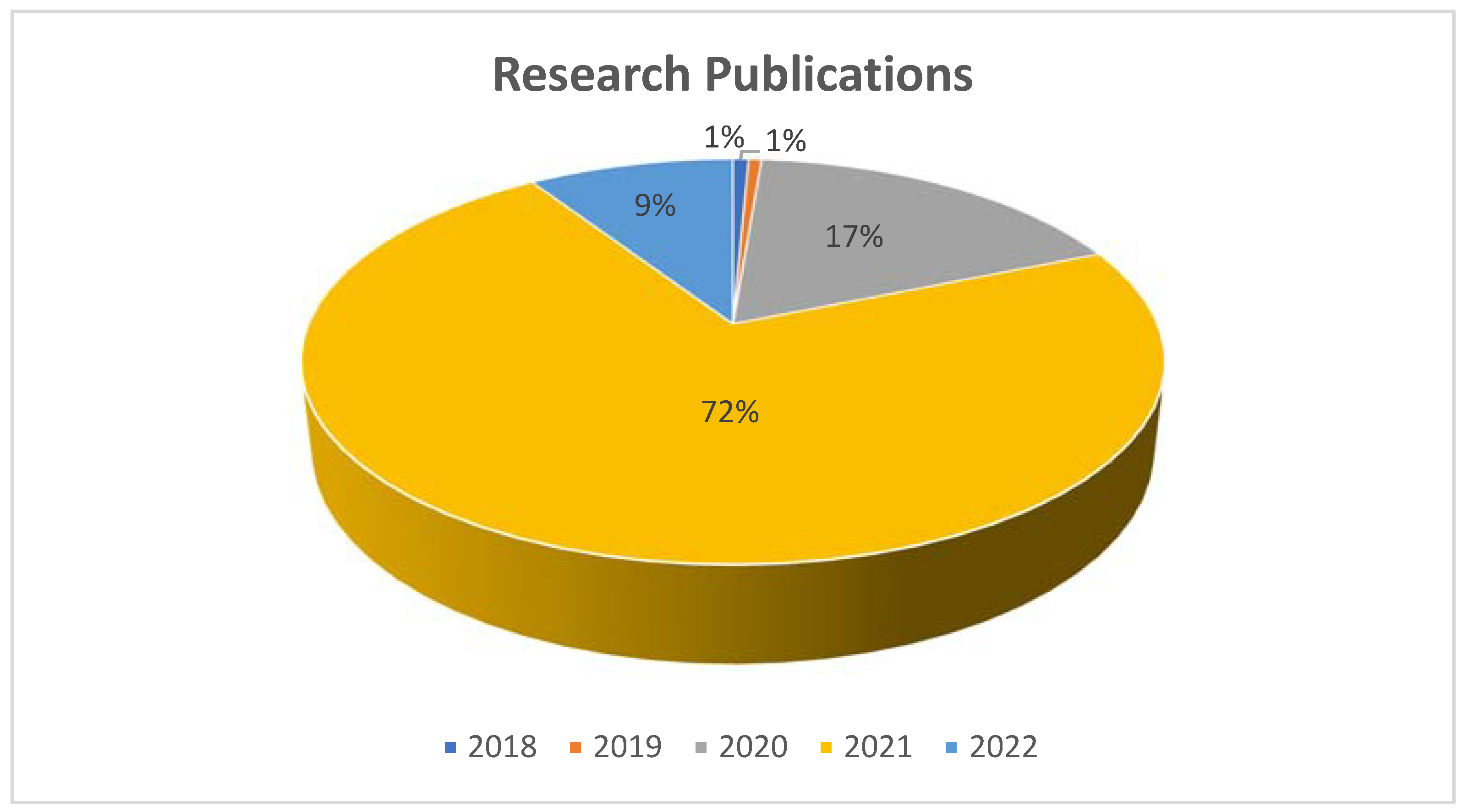
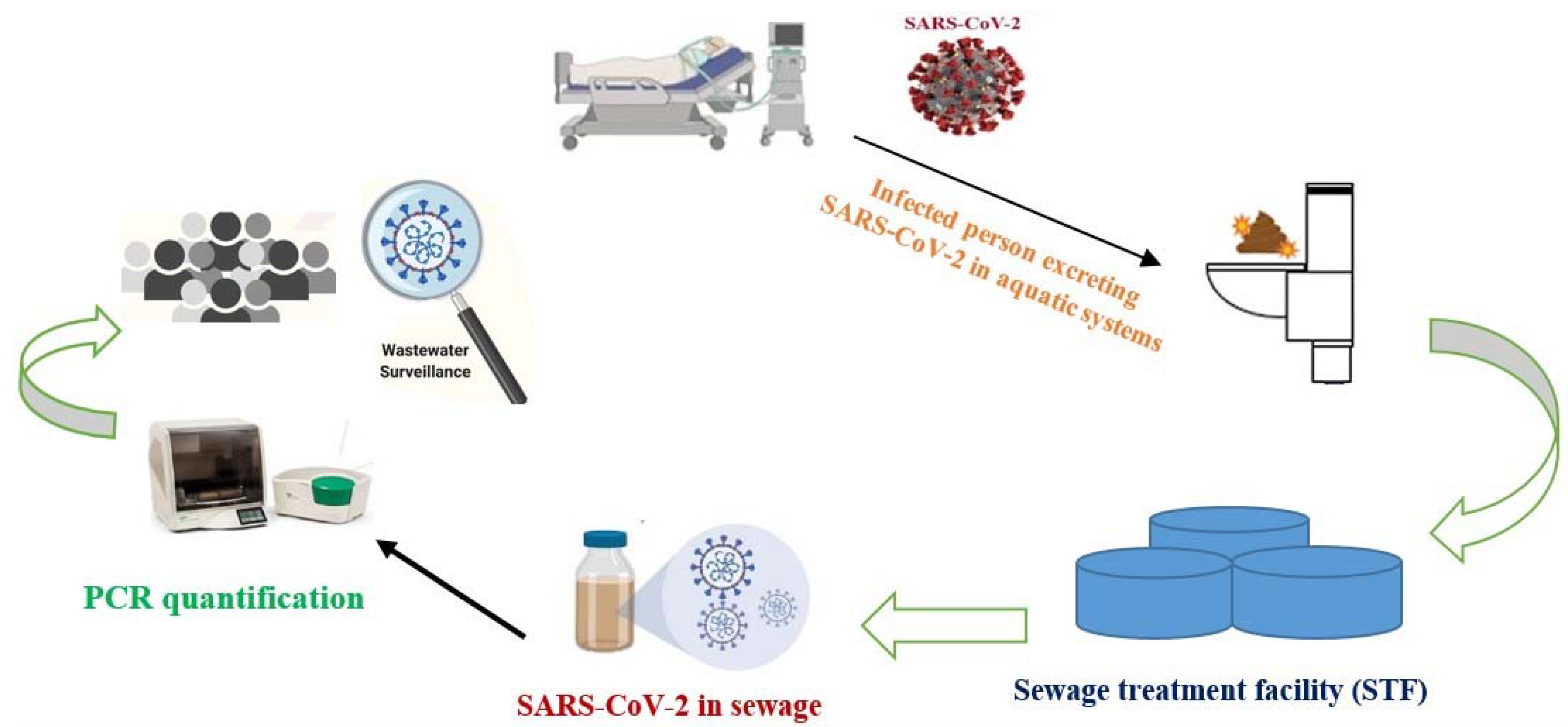

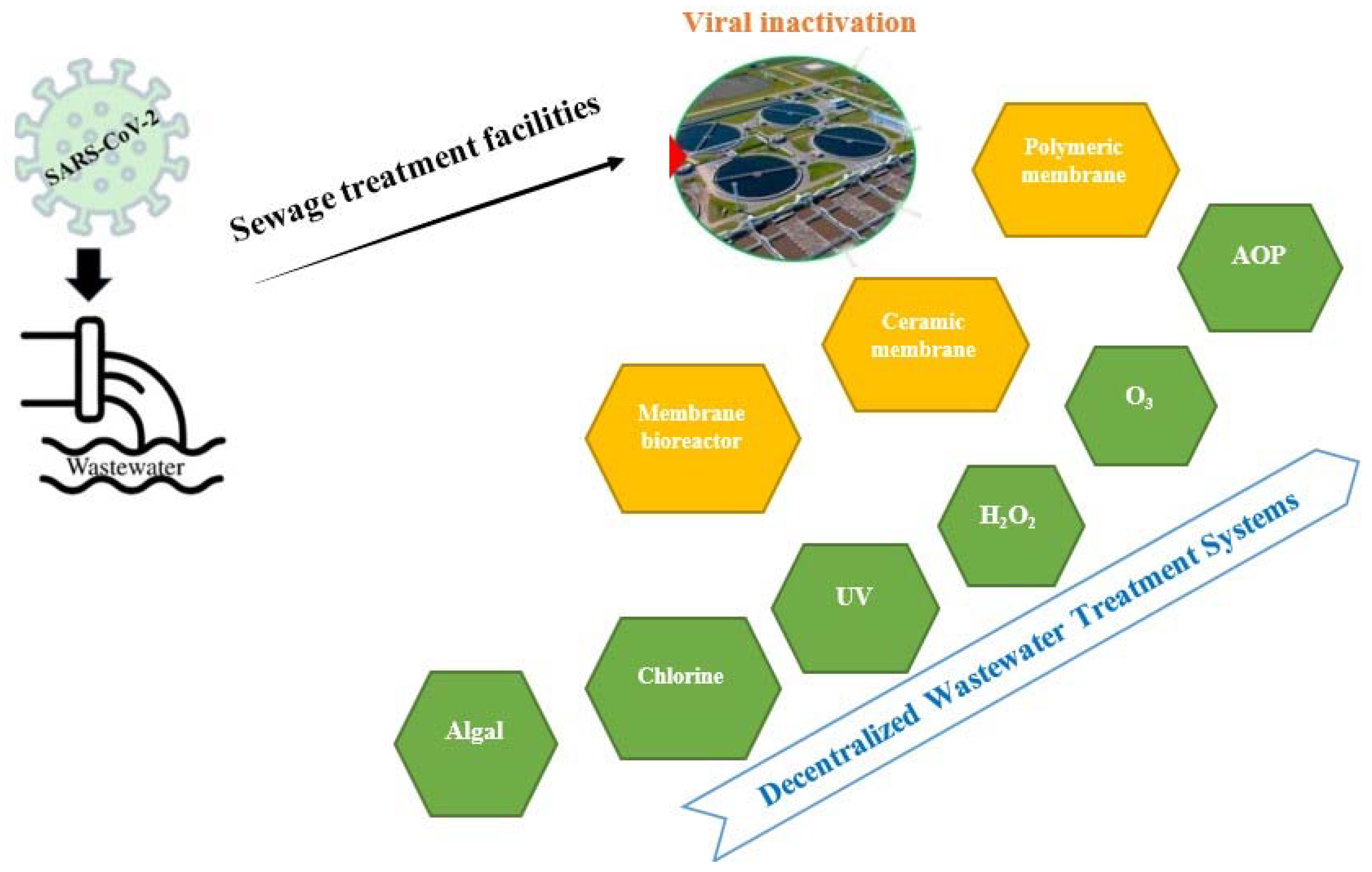
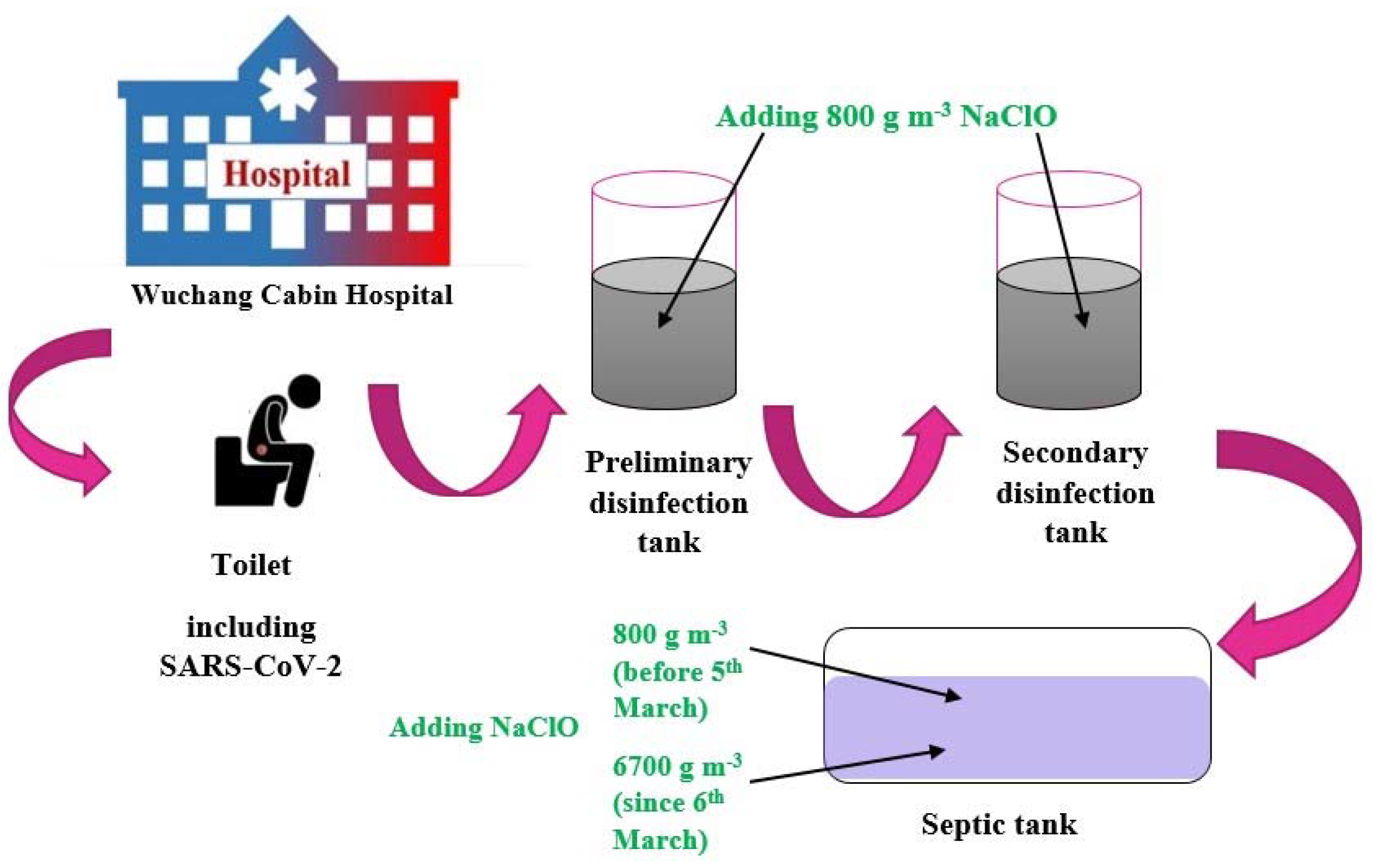
| Location | COVID-19 Prevalence (per 100,000) | SARS-CoV-2 Nucleic Acid Concentration in Wastewater (RNA Copies/mL) | Sources of Sample/Month of Sampling | Total Samples (% Positive) | Genes of SARS CoV-2 Targeted for Detection | Methods for Concentration | Ref. |
|---|---|---|---|---|---|---|---|
| Ourense (Spain) | - | - | WWTP /April 2020 | 5 (100) | N, E, RdRP | Ultrafiltration | [5] |
| Buenos Aires (Argentina) | - | CT value 32–40 | Raw surface water/June–September 2020 | Not Available | N1, N2 | PEG concentration | [27] |
| Pakistan | - | - | Wastewater/March–April 2020 | 78 (27) | ORF 1ab, N | PEG concentration | [28] |
| Calgary (Canada) | - | 0.5 to 11,015.2 | Wastewater/August–December 2020 | 60 (2) | N1, N2, E | 5 μm PVDF filtration, 70% EtOH treatment followed by 4S-silica column for concentration | [29] |
| Toledo (USA) | 9–110.6 | WWTP/July 2020 | 60 (2) | N1 | PEG concentration | [15] | |
| Slovenia | - | 29.65 to 38.12 Cq Up to 10,000 | WWTP/June 2020 | 15 (66.7) | E and RdRP | Ultracentrifugation | [30] |
| Istanbul (Turkey) | - | 9.33 × 104 | WWTP/April 2020 | 9 (77.8) | RdRp | Ultracentrifugation, PEG8000- adsorption electronegative | [31] |
| Netherlands | 0.1–100 | 2 × 103–2.2 × 106 | Untreated wastewater/February–March 2020 | 24 (58.3) | N, E | Ultrafiltration | [32] |
| BneiBrak (Israel) | 366–1001 | - | Untreated wastewater/April 2020 | 26 (38.5) | E-sarbeco | Primary: PEG or Alum; precipitation; secondary: Amicon ultrafiltration | [33] |
| Murcia (Spain) | 8.5–129 | 1 × 105–3.4 × 105 <2.5 × 104 | Untreated Treated wastewater/March–April 2020 | 42 (83.3) 18 (11.1) | N | Aluminum hydroxide adsorption-precipitation | [34] |
| Montpellier (France) | 8 | 1–78 | WWTP/May–July 2020 | - | N1, N3 | Concentration | [35] |
| Czech Republic | 24–561 | Cq 34–40 | WWTP/April–June 2020 | 112 (11.6) | - | Flocculation- centrifugation | [36] |
| Utah (USA) | 2.4–16 | 0.023–1.04 | WWTP/April–May 2020 | 126 (61) | N1, N2 | Centrifuged-electronegative | [20] |
| Wuhan (China) | - | - | WWTP/January 2020 | 4 (0) | ORF 1, N | PEG precipitation of centrifugation supernatant | [26] |
| Milan and Rome (Italy) | - | Not detected | WWTP/February–April 2020 | 12 (50) | ORF1 ab | Polyethylene glycol (PEG) and dextran (DEX) or PEG-dextran | [37] |
| Southern Louisiana (USA) | - | 3.1–7.5 | WWTP/January–April 2020 | 7 (28.6) | ORF 1a, S | Ultrafiltration and adsorption eluting using electronegative membrane | [38] |
| Yamanashi (Japan) | 4.4 | 2.4 | WWTP/ March and May 2020 | 5 (0) | N | Electronegative membrane direction RNA exaction; ultrafiltration | [7] |
| Southeast Queensland (Australia) | - | 1.9 × 101–1.2 × 102 | WWTP/March–April 2020 | 9 (22.2) | N | Electronegative membrane direct RNA exaction; ultrafiltration | [39] |
| Tehran, Qom and Anzali (Iran) | - | - | Treated & untreated wastewater | 24 (58.34) | ORF 1 ab, N | PEG 6000 | [22] |
| Doha (Qatar) | - | 7889 ± 1421– 542,056 ± 25,775 copy/L | WWTP/June–August 2020 | 43 (100) | N | PEG | [24] |
| Pakistan | - | - | WWTP | 78 (26.9) | ORF 1a | PEG/dextran precipitation of centrifuged supernatant | [40] |
| Paris (France) | 0–2000 | 50–3 × 103 | WWTP/March–April 2020 | 23 (100) | RdRP, E | Ultracentrifugation | [25] |
| Ottawa and Gatineau (Canada) | 4.8–57.3 | 1.7–380 | WWTP/April–May 2020 | - | N1, N2 | PEG precipitation | [41] |
| Arizona (USA) | 10.4–993 | Wastewater/August–November 2020 | - | N1, N2 | Ultrafiltration | [18] | |
| Belgrade (Serbia) | - | 5.97 × 103–1.32 × 104 | River water/December 2020 | - | N1, N2, E | Ultracentrifugation | [42] |
| Japan | - | 4.4 × 104 | Untreated wastewater/March–May 2020 | 17 (41.2) | N2, N3, NIID_2019- nCoV_N | PEG precipitation | [43] |
| Metropolitan region (Japan) | - | 0.16–13 | Manhole and WWTP/June–August 2020 | - | N1, N2 | PEG precipitation | [44] |
| Santa Catalina (Brazil) | - | 6.3 × 105 | WWTP/October 2019–March 2020 | - | N1, S, RdRp | PEG precipitation | [45] |
| Ahmedabad (India) | 1000–2700 | 0.78 × 102–8.05 × 102 | Untreated wastewater/May 2020 | 19 (57.89) | ORF1ab, N, S, E | PEG precipitation, Adsorption | [46] |
| Frankfurt (Germany) | - | 4 × 1011–1 × 1015 | WWTP/April | 2 (100) | N, S, ORF 1ab | Electronegative membrane filter | [23] |
| Valencia (Spain) | - | 104–105 | Untreated Treated wastewater/ February–April 2020 | 15 (80) 9 (0) | N | Aluminum flocculation-beef exact precipitation | [47] |
| Niteroi (Brazil) | 51 | 4.9–8.5 | WWTP/April–August 2020 | 12 (41.67) | N1, N2, N3 | Ultracentrifugation | [48] |
| Milan, Turin, and Bologna (Italy) | - | 5.6 × 104 | WWTP/October 2019–February 2020 | 40 (37.5) | ORF1 ab | Dextran and polyethylene glycol, chloroform, centrifugation | [49] |
| Michigan (USA) | - | 104–105 | WWTP/April–May 2020 | 54 (100) | ORF, E, N | NanoCeram filter cartridge | [19] |
| Massachusetts (USA) | 26 | 57–303 | Untreated wastewater/March 2020 | 2 (100) | N1, N2, N3 | Polyethylene glycol-8000 (PEG 8000) | [21] |
| The United Arab Emirates (UAE) | - | 7.50 × 102–3.40 × 104 | Treated & untreated WWTP/May and June 2020 | 36 (77.8) | RdRP | Ultrafiltration columns, and PEG/TRIzol | [50] |
| Disinfection-Based Strategy | Treatment Technology | Crucial Details | Inactivation Ratio | Ref. | |
|---|---|---|---|---|---|
| Chlorine-containing disinfectant | Sodium hypochlorite (NaClO) | 6700 g m−3 | Contact time = 1.5 h V = 60 to 200 m3 | Complete removal | [26] |
| UV inactivation | UV254 | An improved model to predict SARS-CoV-2 inactivation | UV dose of 3 mJ cm2 without attenuation in water | 2-log reduction | [129] |
| AOPs | Effervescent ferrate (VI)-based tablets | Initial concentration of 6400 copy L−1 | Three different tablets: -Pure potassium ferrate of 125 mg -mass ratio 1:2:1 of potassium ferrate: citric acid, anhydrous: sodium hydrogen carbonate -mass ratio 1:4:1 of potassium ferrate: sodium dihydrogen phosphate: sodium hydrogen carbonate | -100% RNA removal -80–100% RNA removal -70–94% RNA removal | [145] |
| AOPs | Electrochemical oxidation | NiOOH as anode catalyst Na2CO3 as electrolyte | -Voltage of 5 V and time of 5 min -Voltage of 5 V and time of 30 s | -99.99% -95% | [143] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paital, B.; Das, K.; Malekdar, F.; Sandoval, M.A.; Niaragh, E.K.; Frontistis, Z.; Behera, T.R.; Balacco, G.; Sangkham, S.; Hati, A.K.; et al. A State-of-the-Art Review on SARS-CoV-2 Virus Removal Using Different Wastewater Treatment Strategies. Environments 2022, 9, 110. https://doi.org/10.3390/environments9090110
Paital B, Das K, Malekdar F, Sandoval MA, Niaragh EK, Frontistis Z, Behera TR, Balacco G, Sangkham S, Hati AK, et al. A State-of-the-Art Review on SARS-CoV-2 Virus Removal Using Different Wastewater Treatment Strategies. Environments. 2022; 9(9):110. https://doi.org/10.3390/environments9090110
Chicago/Turabian StylePaital, Biswaranjan, Kajari Das, Fatemeh Malekdar, Miguel A. Sandoval, Elnaz Karamati Niaragh, Zacharias Frontistis, Tapas Ranjan Behera, Gabriella Balacco, Sarawut Sangkham, Akshaya Kumar Hati, and et al. 2022. "A State-of-the-Art Review on SARS-CoV-2 Virus Removal Using Different Wastewater Treatment Strategies" Environments 9, no. 9: 110. https://doi.org/10.3390/environments9090110








