Effects of Reduced Seawater pH and Oil Contamination on Bacterial Communities and Biochemical Markers of Estuarine Animal Hosts
Abstract
1. Introduction
2. Materials and Methods
2.1. Microcosm Experiment
2.2. DNA Extraction
2.3. Illumina MiSeq Sequencing and Sequence Analysis
2.4. Biochemical Markers Assay
2.5. Data Analysis
2.6. Predictive Functional Profiles
2.7. Denaturing Gradient Gel Electrophoresis (DGGE)
3. Results and Discussion
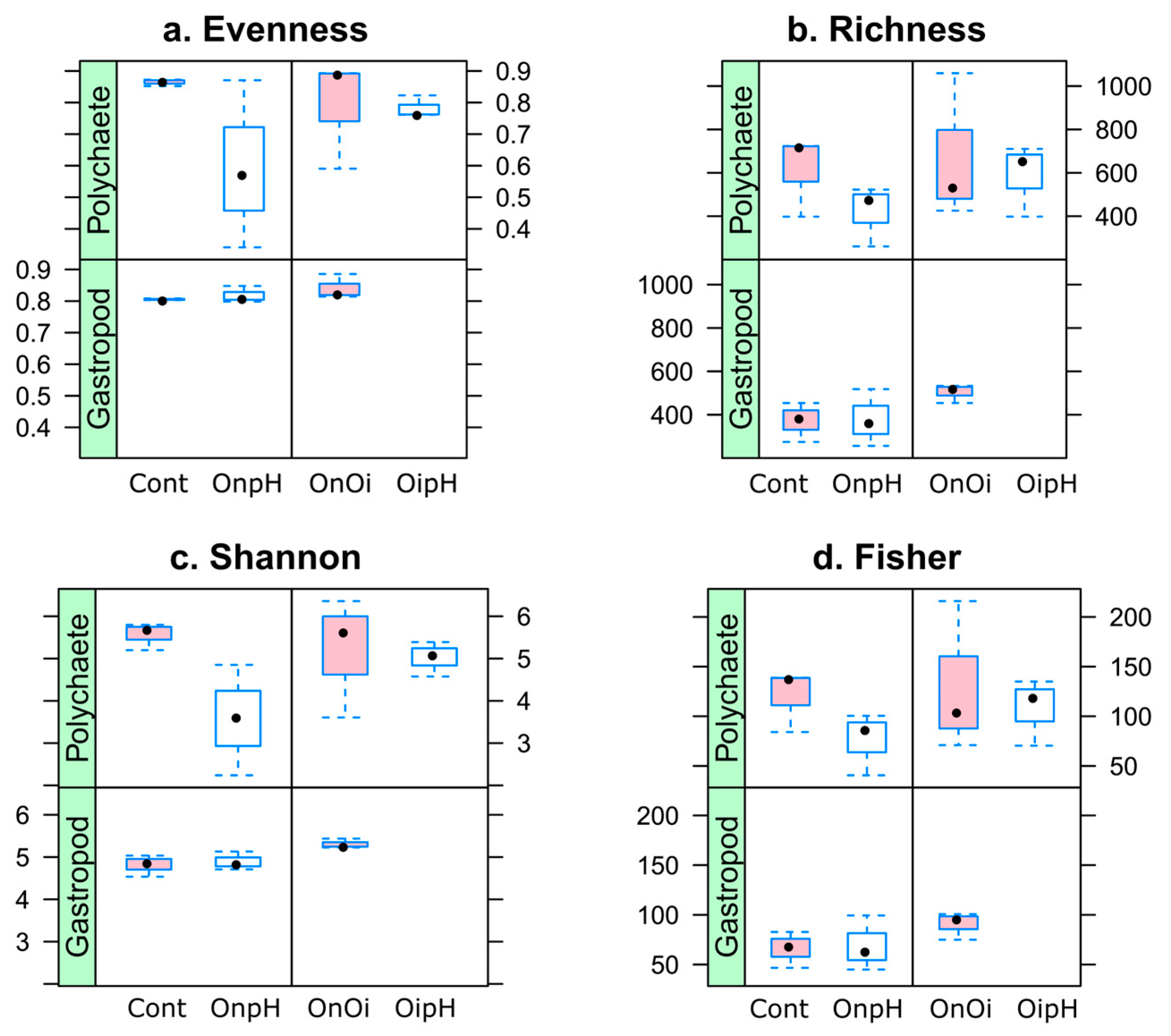
3.1. Diversity and Composition of H. diversicolor- and P. ulvae-Associated Bacterial Communities
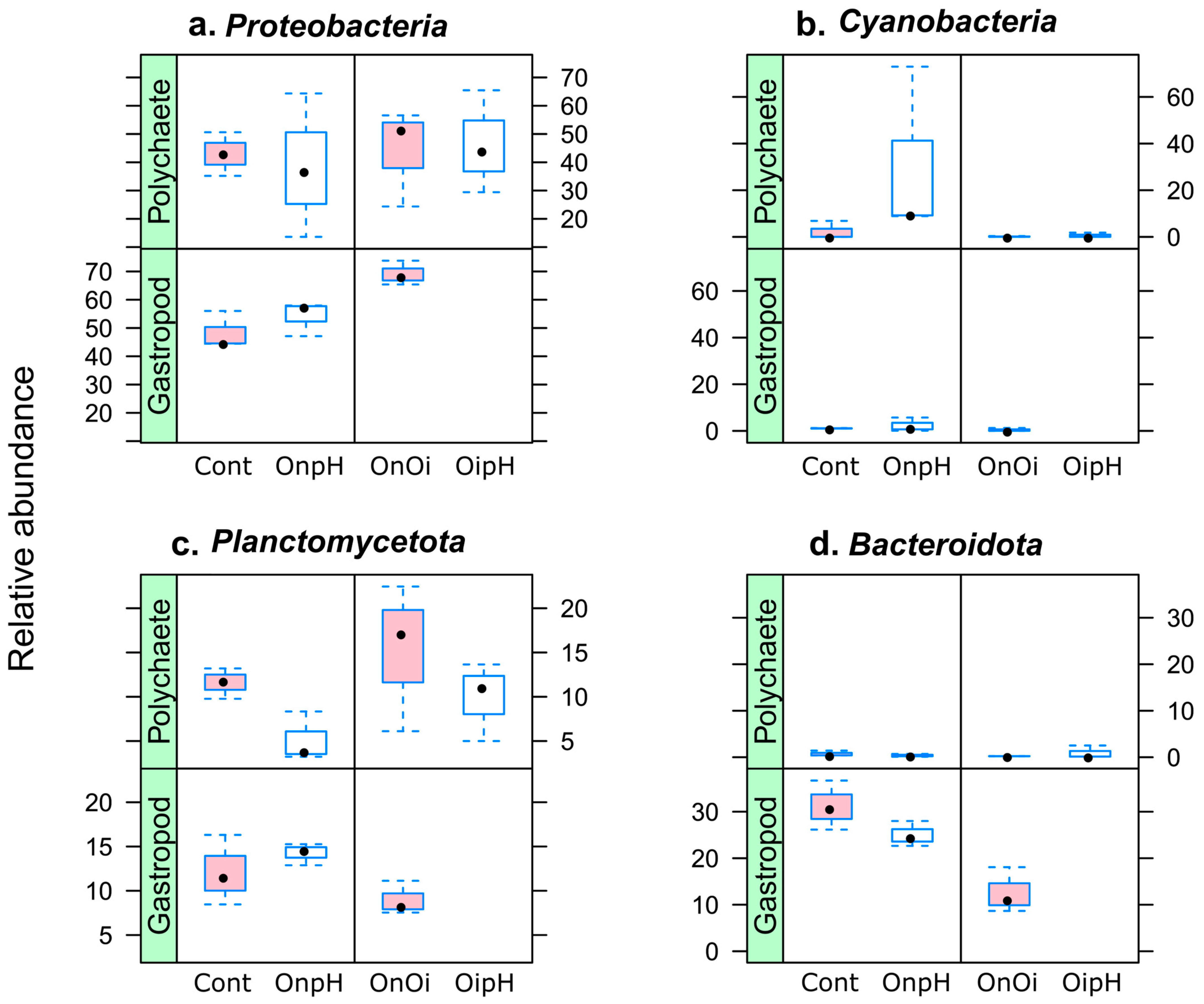
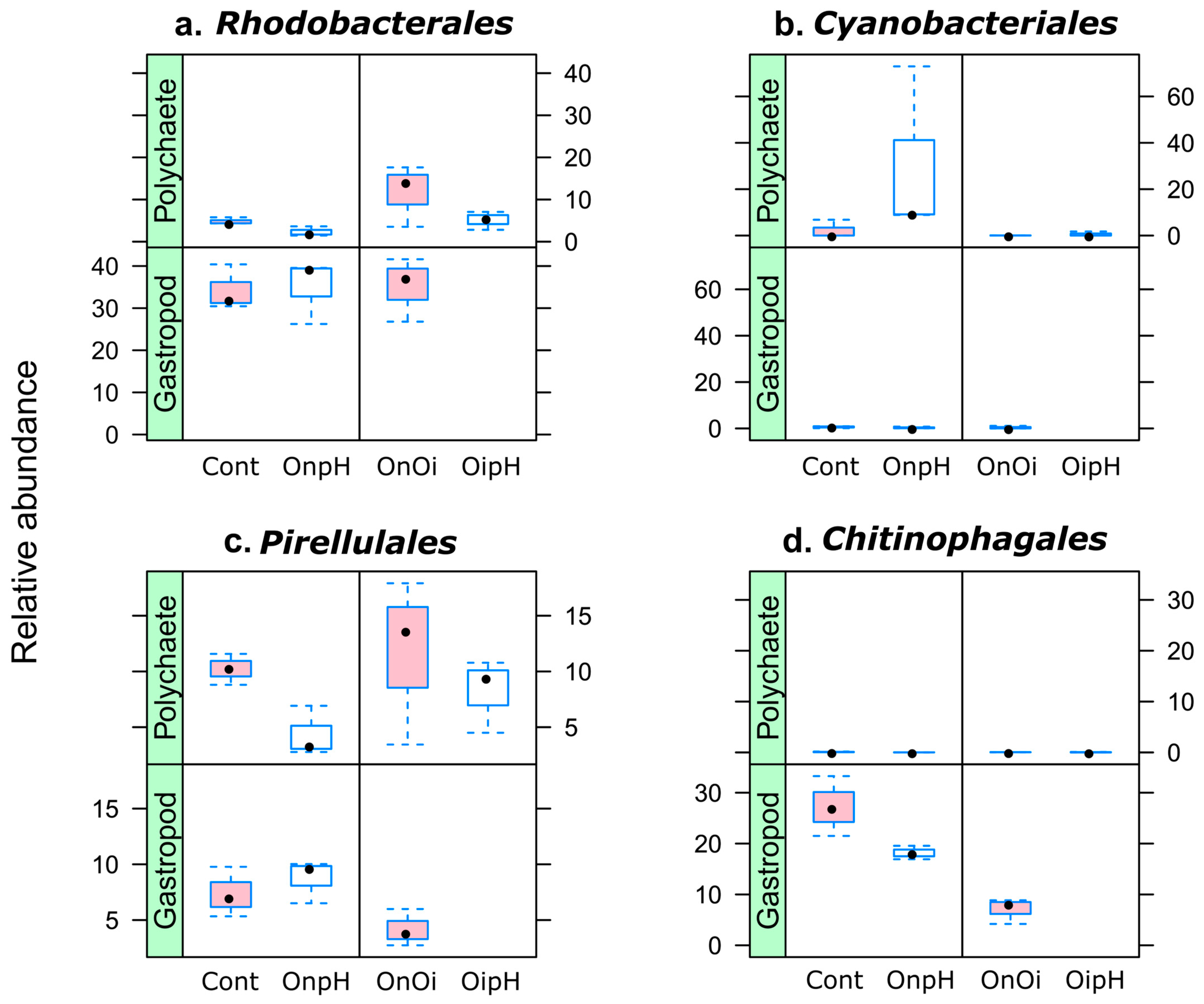
3.2. Taxonomic Composition of H. diversicolor- and P. ulvae-Associated Bacterial Communities
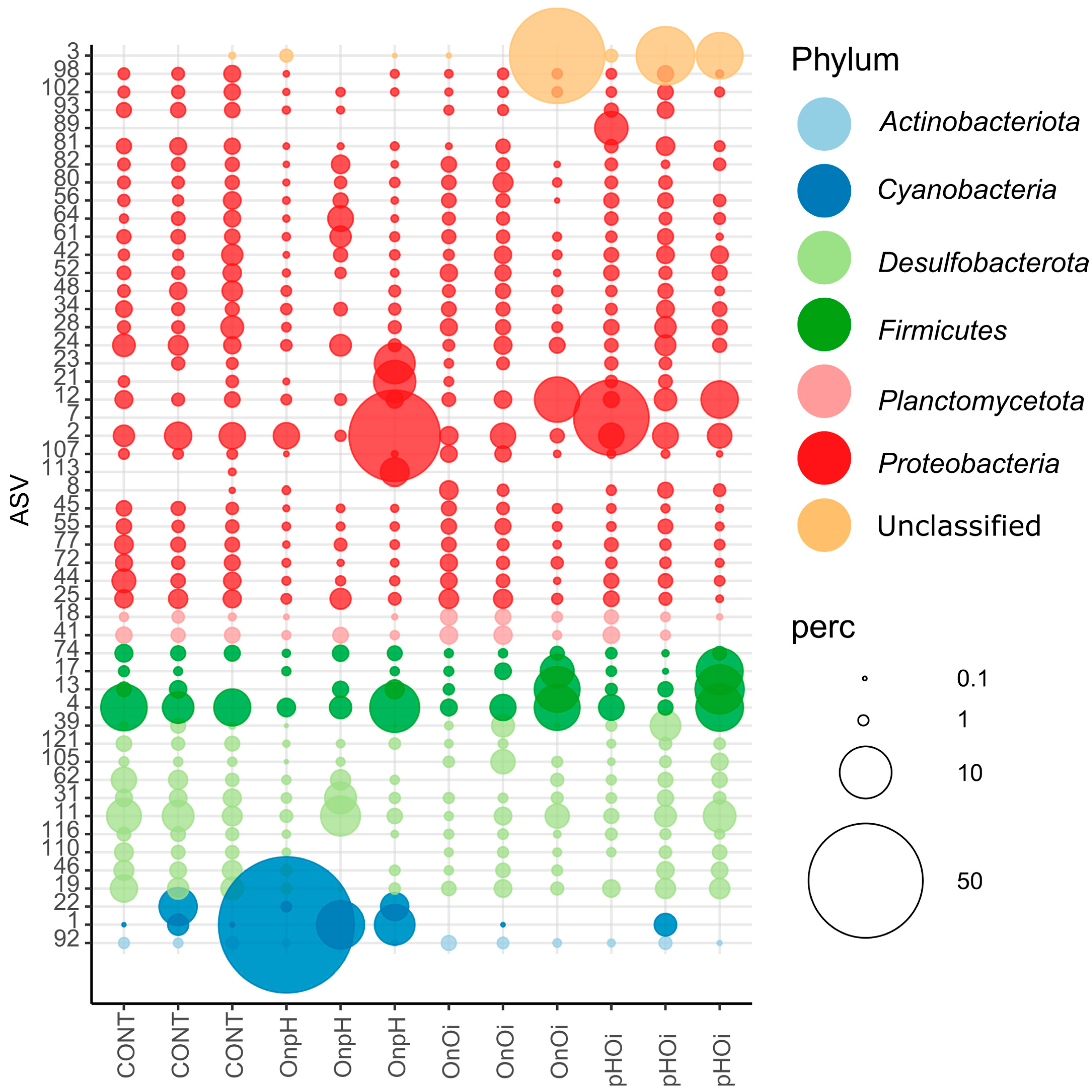
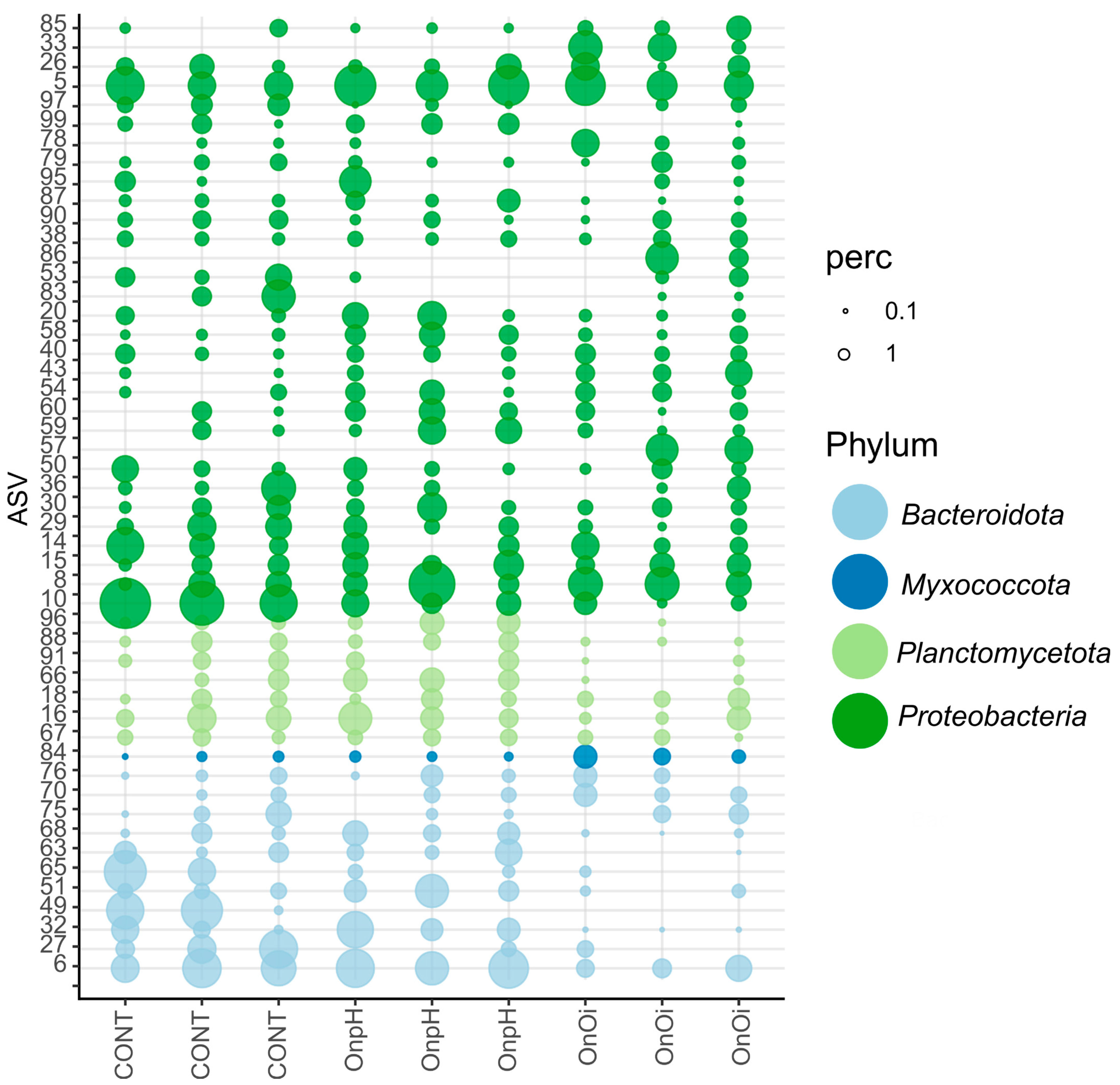
3.3. ASV Compositional Analysis
3.4. Predicted Functional Profiles
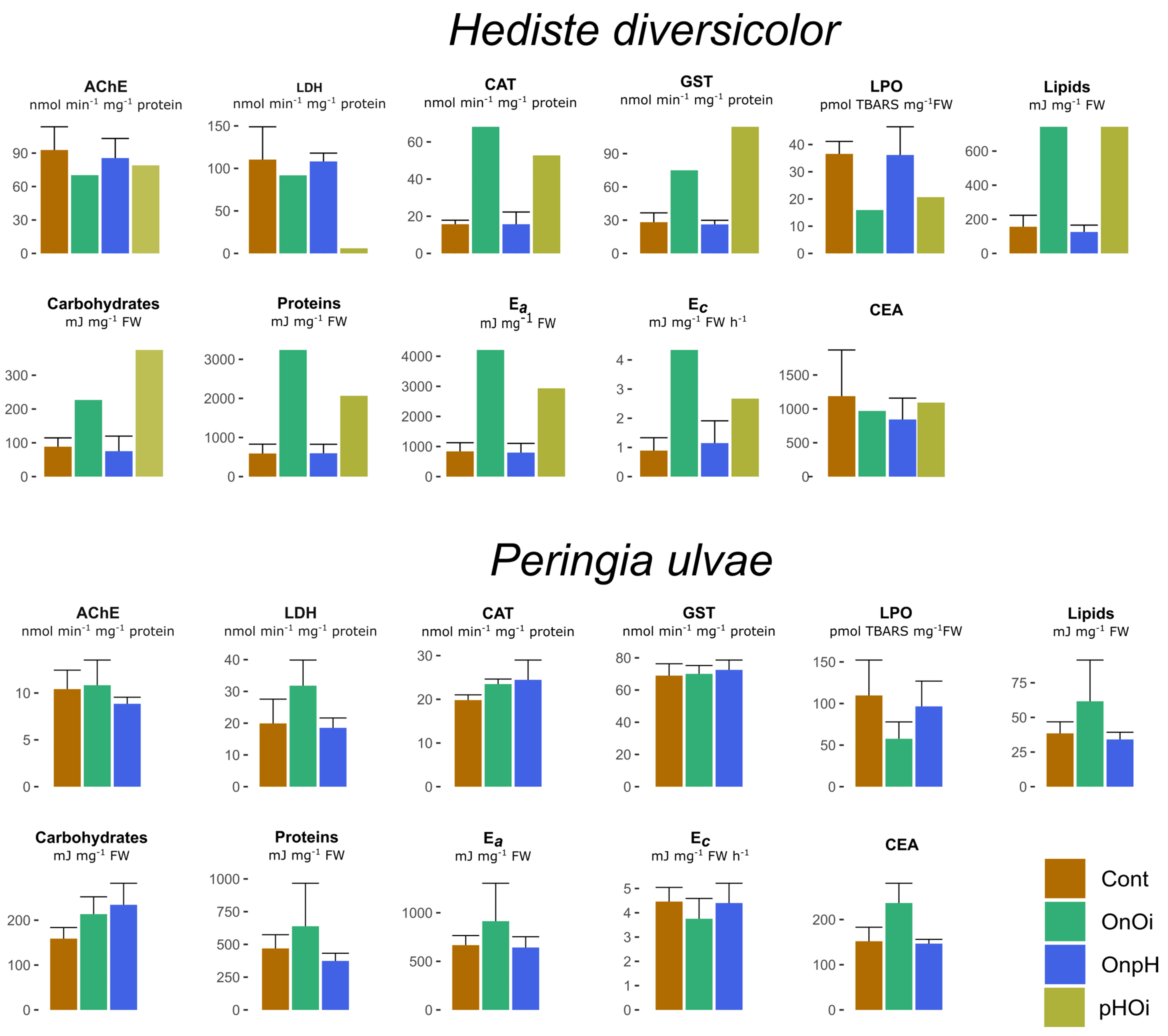
3.5. Biochemical Markers in the Polychaete Hediste diversicolor
3.6. Biochemical Markers in the Gastropod Peringia ulvae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Robinson, C.J.; Bohannan, B.J.M.; Young, V.B. From Structure to Function: The Ecology of Host-Associated Microbial Communities. Microbiol. Mol. Biol. Rev. 2010, 74, 453–476. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, L.G.E.; Leray, M.; O’Dea, A.; Yuen, B.; Peixoto, R.S.; Pereira, T.J.; Bik, H.M.; Coil, D.A.; Duffy, J.E.; Herre, E.A.; et al. Host-Associated Microbiomes Drive Structure and Function of Marine Ecosystems. PLoS Biol. 2019, 17, e3000533. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic Insights into the Marine Sponge Microbiome. Nat Rev Microbiol 2012, 10, 641–654. [Google Scholar] [CrossRef] [PubMed]
- De Goeij, J.M.; Van Oevelen, D.; Vermeij, M.J.A.; Osinga, R.; Middelburg, J.J.; de Goeij, A.F.P.M.; Admiraal, W. Surviving in a Marine Desert: The Sponge Loop Retains Resources within Coral Reefs. Science 2013, 342, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Trevathan-Tackett, S.M.; Sherman, C.D.H.; Huggett, M.J.; Campbell, A.H.; Laverock, B.; Hurtado-McCormick, V.; Seymour, J.R.; Firl, A.; Messer, L.F.; Ainsworth, T.D.; et al. A Horizon Scan of Priorities for Coastal Marine Microbiome Research. Nat. Ecol. Evol. 2019, 3, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S. A, Rosado AS, Bourne DG. 2017. Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 2017, 8, 341. [Google Scholar]
- Coelho, F.J.R.C.; Cleary, D.F.R.; Costa, R.; Ferreira, M.; Polónia, A.R.M.; Silva, A.M.S.; Simões, M.M.Q.; Oliveira, V.; Gomes, N.C.M. Multitaxon Activity Profiling Reveals Differential Microbial Response to Reduced Seawater PH and Oil Pollution. Mol. Ecol. 2016, 25, 4645–4659. [Google Scholar] [CrossRef]
- Louvado, A.; Coelho, F.J.R.C.; Gomes, H.; Cleary, D.F.R.; Cunha, Â.; Gomes, N.C.M. Independent and Interactive Effects of Reduced Seawater PH and Oil Contamination on Subsurface Sediment Bacterial Communities. Environ. Sci. Pollut. Res. 2018, 25, 32756–32766. [Google Scholar] [CrossRef]
- Coelho, F.J.R.C.; Cleary, D.F.R.; Rocha, R.J.M.; Calado, R.; Castanheira, J.M.; Rocha, S.M.; Silva, A.M.S.; Simões, M.M.Q.; Oliveira, V.; Lillebø, A.I. Unraveling the Interactive Effects of Climate Change and Oil Contamination on Laboratory-simulated Estuarine Benthic Communities. Glob. Chang. Biol. 2015, 21, 1871–1886. [Google Scholar] [CrossRef]
- Baltar, F.; Palovaara, J.; Vila-Costa, M.; Salazar, G.; Calvo, E.; Pelejero, C.; Marrasé, C.; Gasol, J.M.; Pinhassi, J. Response of Rare, Common and Abundant Bacterioplankton to Anthropogenic Perturbations in a Mediterranean Coastal Site. FEMS Microbiol. Ecol. 2015, 91, fiv058. [Google Scholar] [CrossRef]
- Friedlingstein, P.; O’sullivan, M.; Jones, M.W.; Andrew, R.M.; Gregor, L.; Hauck, J.; Le Quéré, C.; Luijkx, I.T.; Olsen, A.; Peters, G.P. Global Carbon Budget 2022. Earth Syst. Sci. Data 2022, 14, 4811–4900. [Google Scholar] [CrossRef]
- Doney, S.C.; Busch, D.S.; Cooley, S.R.; Kroeker, K.J. The Impacts of Ocean Acidification on Marine Ecosystems and Reliant Human Communities. Annu. Rev. Environ. Resour. 2020, 45, 83–112. [Google Scholar] [CrossRef]
- Bindoff, N.L.; Cheung, W.W.L.; Kairo, J.G.; Arístegui, J.; Guinder, V.A.; Hallberg, R.; Hilmi, N.; Jiao, N.; Karim, M.S.; Levin, L. Changing Ocean, Marine Ecosystems, and Dependent Communities; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar]
- Caldeira, K.; Wickett, M.E. Anthropogenic Carbon and Ocean PH. Nature 2003, 425, 365. [Google Scholar] [CrossRef]
- Militon, C.; Atkinson, A.; Michotey, V.; Jeziorski, C.; Cravo-Laureau, C.; Duran, R.; Bonin, P.; Cuny, P. Metatranscriptomes of Oil-Contaminated Marine Coastal Sediment Affected by Oil Addition and/or by the Bioturbating Activity of the Marine Polychaete Hediste Diversicolor: Who Are the Microbial Players? Mar. Genom. 2016, 29, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Stauffert, M.; Cravo-Laureau, C.; Duran, R. Dynamic of Sulphate-Reducing Microorganisms in Petroleum-Contaminated Marine Sediments Inhabited by the Polychaete Hediste Diversicolor. Environ. Sci. Pollut. Res. 2015, 22, 15273–15284. [Google Scholar] [CrossRef] [PubMed]
- Dale, H.; Solan, M.; Lam, P.; Cunliffe, M. Sediment Microbial Assemblage Structure Is Modified by Marine Polychaete Gut Passage. FEMS Microbiol. Ecol. 2019, 95, fiz047. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.V.M.; Moreira-Santos, M.; Patrício, J.; Martins, I.; Moreno-Garrido, I.; Blasco, J.; Marques, J.C.; Ribeiro, R. Feeding Niche Preference of the Mudsnail Peringia Ulvae. Mar. Freshw. Res. 2015, 66, 573–581. [Google Scholar] [CrossRef]
- Hale, R.; Mavrogordato, M.N.; Tolhurst, T.J.; Solan, M. Characterizations of How Species Mediate Ecosystem Properties Require More Comprehensive Functional Effect Descriptors. Sci. Rep. 2014, 4, 6463. [Google Scholar] [CrossRef]
- Ulvae, P. Dietary bioavailability of cadmium presented to the gastropod Peringia ulvae as quantum dots and in ionic form. Environ. Toxicol. Chem. 2013, 32, 2621–2629. [Google Scholar]
- Coelho, F.J.R.C.; Santos, A.L.; Coimbra, J.; Almeida, A.; Cunha, Â.; Cleary, D.F.R.; Calado, R.; Gomes, N.C.M. Interactive Effects of Global Climate Change and Pollution on Marine Microbes: The Way Ahead. Ecol. Evol. 2013, 3, 1808–1818. [Google Scholar] [CrossRef]
- Louvado, A.; Coelho, F.J.R.C.; Oliveira, V.; Gomes, H.; Cleary, D.F.R.; Simões, M.M.Q.; Cunha, A.; Gomes, N.C.M. Microcosm Evaluation of the Impact of Oil Contamination and Chemical Dispersant Addition on Bacterial Communities and Sediment Remediation of an Estuarine Port Environment. J. Appl. Microbiol. 2019, 127, 134–149. [Google Scholar] [CrossRef]
- Cravo-Laureau, C.; Jézéquel, R.; Gilbert, F.; Cuny, P.; Duran, R. Mudflat Benthic Spill Simulations BT—Hydrocarbon and Lipid Microbiology Protocols: Meso- and Microcosms; McGenity, T.J., Timmis, K.N., Nogales, B., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2017; pp. 79–89. ISBN 978-3-662-53108-2. [Google Scholar]
- Gattuso, J.-P.; Lavigne, H. Technical Note: Approaches and Software Tools to Investigate the Impact of Ocean Acidification. Biogeosciences 2009, 6, 2121–2133. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 1, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Coelho, F.J.R.C.; Rocha, R.J.M.; Pires, A.C.C.; Ladeiro, B.; Castanheira, J.M.; Costa, R.; Almeida, A.; Cunha, Â.; Lillebø, A.I.; Ribeiro, R. Development and Validation of an Experimental Life Support System for Assessing the Effects of Global Climate Change and Environmental Contamination on Estuarine and Coastal Marine Benthic Communities. Glob. Chang. Biol. 2013, 19, 2584–2595. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.2-1. 2015.
- R. Core Team. R: A Language and Environment for Statistical Computing (Version 2.12. 0); R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Madden, T. The BLAST Sequence Analysis Tool. In The NCBI Handbook; The National Library of Medicine: Bethesda, MD, USA, 2003. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Wieland, G.; Schönfeld, J.; Schönwälder, A.; Gomes, N.C.M.; Smalla, K. Bacterial Community Profiling Using DGGE or TGGE Analysis. Environ. Mol. Microbiol. Protoc. Appl. 2001, 9, 177–190. [Google Scholar]
- Smalla, K.; Wieland, G.; Buchner, A.; Zock, A.; Parzy, J.; Kaiser, S.; Roskot, N.; Heuer, H.; Berg, G. Bulk and Rhizosphere Soil Bacterial Communities Studied by Denaturing Gradient Gel Electrophoresis: Plant-Dependent Enrichment and Seasonal Shifts Revealed. Appl. Environ. Microbiol. 2001, 67, 4742–4751. [Google Scholar] [CrossRef] [PubMed]
- Hochstein, R.; Zhang, Q.; Sadowsky, M.J.; Forbes, V.E. The Deposit Feeder Capitella Teleta Has a Unique and Relatively Complex Microbiome Likely Supporting Its Ability to Degrade Pollutants. Sci. Total Environ. 2019, 670, 547–554. [Google Scholar] [CrossRef]
- Zbinden, M.; Pailleret, M.; Ravaux, J.; Gaudron, S.M.; Hoyoux, C.; Lambourdiere, J.; Waren, A.; Lorion, J.; Halary, S.; Duper-ron, S. Bacterial communities associated with the wood-feeding gastropod Pectinodonta sp. (Patellogastropoda, Mollusca). FEMS 2010, 74, 450–463. [Google Scholar] [CrossRef]
- Chung, H.C.; Lee, O.O.; Huang, Y.L.; Mok, S.Y.; Kolter, R.; Qian, P.Y. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 2010, 4, 817–828. [Google Scholar] [CrossRef]
- McIlroy, S.J.; Nielsen, P.H. The Family Saprospiraceae BT—The Prokaryotes: Other Major Lineages of Bacteria and The Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 863–889. ISBN 978-3-642-38954-2. [Google Scholar]
- Kondrotaite, Z.; Valk, L.C.; Petriglieri, F.; Singleton, C.; Nierychlo, M.; Dueholm, M.K.D.; Nielsen, P.H. Diversity and Ecophysiology of the Genus OLB8 and Other Abundant Uncultured Saprospiraceae Genera in Global Wastewater Treatment Systems. Front. Microbiol. 2022, 13, 917553. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, Q.; Huang, Q.; Wang, S.; Qin, X.; Ren, T.; Xie, R.; Su, H. Significant Shifts in Microbial Communities Associated with Scleractinian Corals in Response to Algae Overgrowth. Microorganisms 2022, 10, 2196. [Google Scholar] [CrossRef]
- Sun, L.; Gao, P.; Li, Y.; Wang, C.; Ding, N.; Chen, J.; Song, Y.; Liu, C.; Song, L.; Wang, R. Microbial Community Coexisting with Harmful Alga Karenia Mikimotoi and Microbial Control of Algal Bloom in Laboratory. J. Oceanol. Limnol. 2022, 40, 1027–1038. [Google Scholar] [CrossRef]
- Bowman, J.P.; Nowak, B. Salmonid Gill Bacteria and Their Relationship to Amoebic Gill Disease. J. Fish. Dis. 2004, 27, 483–492. [Google Scholar] [CrossRef]
- Vijayan, N.; Lema, K.A.; Nedved, B.T.; Hadfield, M.G. Microbiomes of the Polychaete Hydroides Elegans (Polychaeta: Serpulidae) across Its Life-History Stages. Mar. Biol. 2019, 166, 19. [Google Scholar] [CrossRef]
- Neave, M.J.; Michell, C.T.; Apprill, A.; Voolstra, C.R. Endozoicomonas Genomes Reveal Functional Adaptation and Plasticity in Bacterial Strains Symbiotically Associated with Diverse Marine Hosts. Sci. Rep. 2017, 7, 40579. [Google Scholar] [CrossRef] [PubMed]
- Michas, A.; Vestergaard, G.; Trautwein, K.; Avramidis, P.; Hatzinikolaou, D.G.; Vorgias, C.E.; Wilkes, H.; Rabus, R.; Schloter, M.; Schöler, A. More than 2500 Years of Oil Exposure Shape Sediment Microbiomes with the Potential for Syntrophic Degradation of Hydrocarbons Linked to Methanogenesis. Microbiome 2017, 5, 118. [Google Scholar] [CrossRef]
- Wu, X.-L.; Yu, S.-L.; Gu, J.; Zhao, G.-F.; Chi, C.-Q. Filomicrobium Insigne Sp. Nov., Isolated from an Oil-Polluted Saline Soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Golyshin, P.N.; Lang, S.; Moore, E.R.B.; Abraham, W.-R.; Lunsdorf, H.; Timmis, K.N. Alcanivorax Borkumensis Gen. Nov., Sp. Nov., a New, Hydrocarbon-Degrading and Surfactant-Producing Marine Bacterium. Int. J. Syst. Bacteriol. 1998, 48, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.M.; Bertran, E.; Johnston, D.T. Expanded Genomic Sampling Refines Current Understanding of the Distribution and Evolution of Sulfur Metabolisms in the Desulfobulbales. Front. Microbiol. 2021, 12, 666052. [Google Scholar] [CrossRef]
- Thompson, A.W.; Foster, R.A.; Krupke, A.; Carter, B.J.; Musat, N.; Vaulot, D.; Kuypers, M.M.M.; Zehr, J.P. Unicellular Cyanobacterium Symbiotic with a Single-Celled Eukaryotic Alga. Science 2012, 337, 1546–1550. [Google Scholar] [CrossRef]
- Puente-Sánchez, F.; Arce-Rodríguez, A.; Oggerin, M.; García-Villadangos, M.; Moreno-Paz, M.; Blanco, Y.; Rodríguez, N.; Bird, L.; Lincoln, S.A.; Tornos, F.; et al. Viable Cyanobacteria in the Deep Continental Subsurface. Proc. Natl. Acad. Sci. 2018, 115, 10702–10707. [Google Scholar] [CrossRef]
- Thacker, R.W. Impacts of Shading on Sponge-Cyanobacteria Symbioses: A Comparison between Host-Specific and Generalist Associations1. Integr. Comp. Biol. 2005, 45, 369–376. [Google Scholar] [CrossRef]
- Carpenter, E.J.; Foster, R.A. Marine Cyanobacterial Symbioses. In Cyanobacteria in symbiosis; Springer: Berlin/Heidelberg, Germany, 2002; pp. 11–17. [Google Scholar]
- Wang, D.; Ren, J.; Tan, Z.; You, J. Gut Microbial Profiles in Nereis Succinea and Their Contribution to the Degradation of Organic Pollutants. Environ. Sci. Technol. 2020, 54, 6235–6243. [Google Scholar] [CrossRef]
- Sandrini, G.; Ji, X.; Verspagen, J.M.H.; Tann, R.P.; Slot, P.C.; Luimstra, V.M.; Schuurmans, J.M.; Matthijs, H.C.P.; Huisman, J. Rapid Adaptation of Harmful Cyanobacteria to Rising CO2. Proc. Natl. Acad. Sci. 2016, 113, 9315–9320. [Google Scholar] [CrossRef]
- Morrow, K.M.; Bourne, D.G.; Humphrey, C.; Botté, E.S.; Laffy, P.; Zaneveld, J.; Uthicke, S.; Fabricius, K.E.; Webster, N.S. Natural Volcanic CO2 Seeps Reveal Future Trajectories for Host–Microbial Associations in Corals and Sponges. ISME J. 2015, 9, 894–908. [Google Scholar] [CrossRef]
- Stauffert, M.; Cravo-Laureau, C.; Jézéquel, R.; Barantal, S.; Cuny, P.; Gilbert, F.; Cagnon, C.; Militon, C.; Amouroux, D.; Mahdaoui, F.; et al. Impact of Oil on Bacterial Community Structure in Bioturbated Sediments. PLoS ONE 2013, 8, e65347. [Google Scholar] [CrossRef] [PubMed]
- Martins dos Santos, V.; Sabirova, J.; Timmis, K.N.; Yakimov, M.M.; Golyshin, P.N. Alcanivorax Borkumensis BT—Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1265–1288. ISBN 978-3-540-77587-4. [Google Scholar]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate Oil-Degrading Marine Bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, T.N.; Bargiela, R.; Toshchakov, S.V.; Shivaraman, V.; Lunev, E.A.; Yakimov, M.M.; Thomas, D.N.; Golyshin, P.N. Hydrocarbon-Degrading Bacteria Alcanivorax and Marinobacter Associated With Microalgae Pavlova Lutheri and Nannochloropsis Oculata. Front. Microbiol. 2020, 11, 2650. [Google Scholar] [CrossRef] [PubMed]
- Bolch, C.J.S.; Bejoy, T.A.; Green, D.H. Bacterial Associates Modify Growth Dynamics of the Dinoflagellate Gymnodinium Catenatum. Front. Microbiol. 2017, 8, 670. [Google Scholar] [CrossRef] [PubMed]
- Green, D.H.; Llewellyn, L.E.; Negri, A.P.; Blackburn, S.I.; Bolch, C.J.S. Phylogenetic and Functional Diversity of the Cultivable Bacterial Community Associated with the Paralytic Shellfish Poisoning Dinoflagellate Gymnodinium Catenatum. FEMS Microbiol. Ecol. 2004, 47, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Green, D.H.; Hart, M.C.; Küpper, F.C.; Sunda, W.G.; Carrano, C.J. Photolysis of Iron–Siderophore Chelates Promotes Bacterial–Algal Mutualism. Proc. Natl. Acad. Sci. 2009, 106, 17071–17076. [Google Scholar] [CrossRef]
- Shaikhulova, S.; Fakhrullina, G.; Nigamatzyanova, L.; Akhatova, F.; Fakhrullin, R. Worms Eat Oil: Alcanivorax Borkumensis Hydrocarbonoclastic Bacteria Colonise Caenorhabditis Elegans Nematodes Intestines as a First Step towards Oil Spills Zooremediation. Sci. Total Environ. 2021, 761, 143209. [Google Scholar] [CrossRef]
- Pal, S.; Kundu, A.; Banerjee, T.D.; Mohapatra, B.; Roy, A.; Manna, R.; Sar, P.; Kazy, S.K. Genome Analysis of Crude Oil Degrading Franconibacter Pulveris Strain DJ34 Revealed Its Genetic Basis for Hydrocarbon Degradation and Survival in Oil Contaminated Environment. Genomics 2017, 109, 374–382. [Google Scholar] [CrossRef]
- Sierra-García, I.N.; Correa Alvarez, J.; Pantaroto de Vasconcellos, S.; Pereira de Souza, A.; dos Santos Neto, E.V.; de Oliveira, V.M. New Hydrocarbon Degradation Pathways in the Microbial Metagenome from Brazilian Petroleum Reservoirs. PLoS ONE 2014, 9, e90087. [Google Scholar] [CrossRef]
- Bouraoui, Z.; Banni, M.; Ghedira, J.; Clerandeau, C.; Narbonne, J.F.; Boussetta, H. Evaluation of Enzymatic Biomarkers and Lipoperoxidation Level in Hediste Diversicolor Exposed to Copper and Benzo [a] Pyrene. Ecotoxicol. Environ. Saf. 2009, 72, 1893–1898. [Google Scholar] [CrossRef]
- Rodrigues, A.P.; Lehtonen, K.K.; Guilhermino, L.; Guimarães, L. Exposure of Carcinus Maenas to Waterborne Fluoranthene: Accumulation and Multibiomarker Responses. Sci. Total Environ. 2013, 443, 454–463. [Google Scholar] [CrossRef]
- Olivares-Rubio, H.F.; Espinosa-Aguirre, J.J. Acetylcholinesterase Activity in Fish Species Exposed to Crude Oil Hydrocarbons: A Review and New Perspectives. Chemosphere 2021, 264, 128401. [Google Scholar] [CrossRef]
- Ansaldo, M.; Najle, R.; Luquet, C.M. Oxidative Stress Generated by Diesel Seawater Contamination in the Digestive Gland of the Antarctic Limpet Nacella Concinna. Mar. Environ. Res. 2005, 59, 381–390. [Google Scholar] [CrossRef]
- Sazykin, I.S.; Sazykina, M.A.; Khmelevtsova, L.E.; Seliverstova, E.Y.; Karchava, K.S.; Zhuravleva, M. V Antioxidant Enzymes and Reactive Oxygen Species Level of the Achromobacter Xylosoxidans Bacteria during Hydrocarbons Biotransformation. Arch. Microbiol. 2018, 200, 1057–1065. [Google Scholar] [CrossRef]
- Solé, M.; Kopecka-Pilarczyk, J.; Blasco, J. Pollution Biomarkers in Two Estuarine Invertebrates, Nereis Diversicolor and Scrobicularia Plana, from a Marsh Ecosystem in SW Spain. Environ. Int. 2009, 35, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Moreira, S.M.; Lima, I.; Ribeiro, R.; Guilhermino, L. Effects of Estuarine Sediment Contamination on Feeding and on Key Physiological Functions of the Polychaete Hediste Diversicolor: Laboratory and in Situ Assays. Aquat. Toxicol. 2006, 78, 186–201. [Google Scholar] [CrossRef]
- Nechev, J.T.; Khotimchenko, S.V.; Ivanova, A.P.; Stefanov, K.L.; Dimitrova-Konaklieva, S.D.; Andreev, S.; Popov, S.S. Effect of Diesel Fuel Pollution on the Lipid Composition of Some Wide-Spread Black Sea Algae and Invertebrates. Z. Für Naturforschung C 2002, 57, 1–5. [Google Scholar] [CrossRef]
- Carvalho, R.N.; Burchardt, A.D.; Sena, F.; Mariani, G.; Mueller, A.; Bopp, S.K.; Umlauf, G.; Lettieri, T. Gene Biomarkers in Diatom Thalassiosira Pseudonana Exposed to Polycyclic Aromatic Hydrocarbons from Contaminated Marine Surface Sediments. Aquat. Toxicol. 2011, 101, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Grilo, T.F.; Lopes, A.R.; Sampaio, E.; Rosa, R.; Cardoso, P.G. Sex Differences in Oxidative Stress Responses of Tropical Topshells (Trochus Histrio) to Increased Temperature and High PCO2. Mar. Pollut. Bull. 2018, 131, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Boldina-Cosqueric, I.; Amiard, J.-C.; Amiard-Triquet, C.; Dedourge-Geffard, O.; Métais, I.; Mouneyrac, C.; Moutel, B.; Berthet, B. Biochemical, Physiological and Behavioural Markers in the Endobenthic Bivalve Scrobicularia Plana as Tools for the Assessment of Estuarine Sediment Quality. Ecotoxicol. Environ. Saf. 2010, 73, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Hu, M.; Shang, Y.; Wu, F.; Huang, X.; Dupont, S.; Storch, D.; Pörtner, H.-O.; Li, J.; Lu, W. Antioxidant Response of the Hard Shelled Mussel Mytilus Coruscus Exposed to Reduced PH and Oxygen Concentration. Ecotoxicol. Environ. Saf. 2017, 137, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Thangal, S.H.; Nivetha, M.; Muttharasi, C.; Anandhan, K.; Muralisankar, T. Effects of Acidified Seawater on Biological and Physiological Responses of Artemia Franciscana. Mar. Pollut. Bull. 2021, 169, 112476. [Google Scholar] [CrossRef] [PubMed]
- Freitas, R.; Pires, A.; Moreira, A.; Wrona, F.J.; Figueira, E.; Soares, A.M.V.M. Biochemical Alterations Induced in Hediste Diversicolor under Seawater Acidification Conditions. Mar. Environ. Res. 2016, 117, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Jeong, C.-B.; Wang, M.; Hagiwara, A.; Lee, J.-S. Transgenerational Acclimation to Changes in Ocean Acidification in Marine Invertebrates. Mar. Pollut. Bull. 2020, 153, 111006. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.M.; Torres, R.; Acuña, K.; Duarte, C.; Manriquez, P.H.; Lardies, M.; Lagos, N.A.; Vargas, C.; Aguilera, V. Impact of Medium-Term Exposure to Elevated PCO2 Levels on the Physiological Energetics of the Mussel Mytilus Chilensis. Chemosphere 2013, 90, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Michaelidis, B.; Ouzounis, C.; Paleras, A.; Pörtner, H.O. Effects of Long-Term Moderate Hypercapnia on Acid–Base Balance and Growth Rate in Marine Mussels Mytilus Galloprovincialis. Mar. Ecol. Prog. Ser. 2005, 293, 109–118. [Google Scholar] [CrossRef]
- Freitas, R.; De Marchi, L.; Bastos, M.; Moreira, A.; Velez, C.; Chiesa, S.; Wrona, F.J.; Figueira, E.; Soares, A.M.V.M. Effects of Seawater Acidification and Salinity Alterations on Metabolic, Osmoregulation and Oxidative Stress Markers in Mytilus Galloprovincialis. Ecol. Indic. 2017, 79, 54–62. [Google Scholar] [CrossRef]
- Silva, C.S.E.; Novais, S.C.; Lemos, M.F.L.; Mendes, S.; Oliveira, A.P.; Gonçalves, E.J.; Faria, A.M. Effects of Ocean Acidification on the Swimming Ability, Development and Biochemical Responses of Sand Smelt Larvae. Sci. Total Environ. 2016, 563, 89–98. [Google Scholar] [CrossRef]
- Sokołowski, A.; Brulińska, D. The Effects of Low Seawater PH on Energy Storage and Heat Shock Protein 70 Expression in a Bivalve Limecola Balthica. Mar. Environ. Res. 2018, 140, 289–298. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Clairborne, A. Catalase activity. In Handbook of Method for Oxygen Radical Research; Greenwald, R.A., Ed.; CRC Press: Boca Raton, FL, USA, 1985. [Google Scholar]
- De Coen, W.M.; Janssen, C.R. The missing biomarker link: Relationships between effects on the cellular energy allocation biomarker of toxicant-stressed Daphnia magna and corresponding population characteristics. Environ. Toxicol. Chem. Int. J. 2003, 22, 1632–1641. [Google Scholar]
- Diamantino, T.C.; Almeida, E.; Soares, A.M.V.M.; Guilhermino, L. Lactate dehydrogenase activity as an effect criterion in toxicity tests with Daphnia magna straus. Chemosphere 2001, 45, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Dunn, P.K. Tweedie: Evaluation of Tweedie Exponential Family Models, R Packag. Version 2. 2017.
- Ellman, G.L.; Courtney, K.D.; Andres Jr, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases: The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Vassault, A. Lactate dehydrogenase, UV-method with pyruvate and NADH. Methods Enzym. Anal. 1983, 3, 118. [Google Scholar]
- Verslycke, T.; Vercauteren, J.; Devos, C.; Moens, L.; Sandra, P.; Janssen, C.R. Cellular energy allocation in the estuarine mysid shrimp Neomysis integer (Crustacea: Mysidacea) following tributyltin exposure. J. Exp. Mar. Bio. Ecol. 2003, 288, 167–179. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Louvado, A.; Galhano, V.; Lima, F.; Cleary, D.F.R.; Lopes, I.; Gomes, N.C.M.; Coelho, F.J.R.C. Effects of Reduced Seawater pH and Oil Contamination on Bacterial Communities and Biochemical Markers of Estuarine Animal Hosts. Environments 2024, 11, 37. https://doi.org/10.3390/environments11020037
Louvado A, Galhano V, Lima F, Cleary DFR, Lopes I, Gomes NCM, Coelho FJRC. Effects of Reduced Seawater pH and Oil Contamination on Bacterial Communities and Biochemical Markers of Estuarine Animal Hosts. Environments. 2024; 11(2):37. https://doi.org/10.3390/environments11020037
Chicago/Turabian StyleLouvado, António, Victor Galhano, Fernanda Lima, Daniel F. R. Cleary, Isabel Lopes, Newton C. M. Gomes, and Francisco J. R. C. Coelho. 2024. "Effects of Reduced Seawater pH and Oil Contamination on Bacterial Communities and Biochemical Markers of Estuarine Animal Hosts" Environments 11, no. 2: 37. https://doi.org/10.3390/environments11020037
APA StyleLouvado, A., Galhano, V., Lima, F., Cleary, D. F. R., Lopes, I., Gomes, N. C. M., & Coelho, F. J. R. C. (2024). Effects of Reduced Seawater pH and Oil Contamination on Bacterial Communities and Biochemical Markers of Estuarine Animal Hosts. Environments, 11(2), 37. https://doi.org/10.3390/environments11020037








