Different Responses of Microbiota across Intestinal Tract to Enterococcus faecium HDRsEf1 and Their Correlation with Inflammation in Weaned Piglets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry
2.2. Slaughtering and Sampling
2.3. DNA Extraction, 16S rRNA Gene Amplification, and High-Through Sequencing
2.4. Analysis of Sequencing Data
2.5. RNA Extraction and RT-qPCR of Small Intestine
2.6. Morphological Analysis of Small Intestine
2.7. Microbial Genomic DNA Extraction and Quantification
2.8. Analysis of Serum Inflammatory Factors
2.9. Statistical Analysis
3. Results
3.1. High-Throughput Sequencing Data
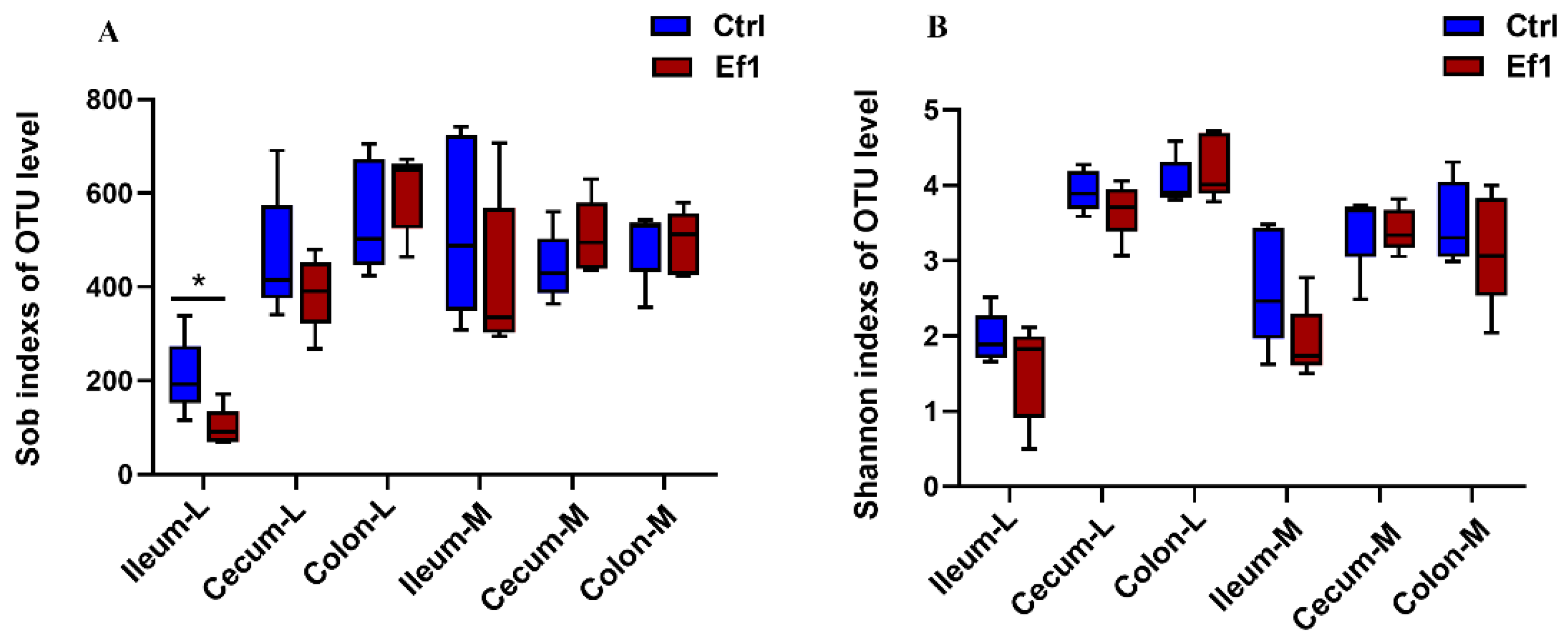
3.2. Effects of HDRsEf1 on Microbial Alpha Diversity in Different Niches
3.3. Effects of HDRsEf1 on Total Bacterial Load in Different Gut Niches
3.4. Effects of HDRsEf1 on Microbial Beta Diversity in Different Niches
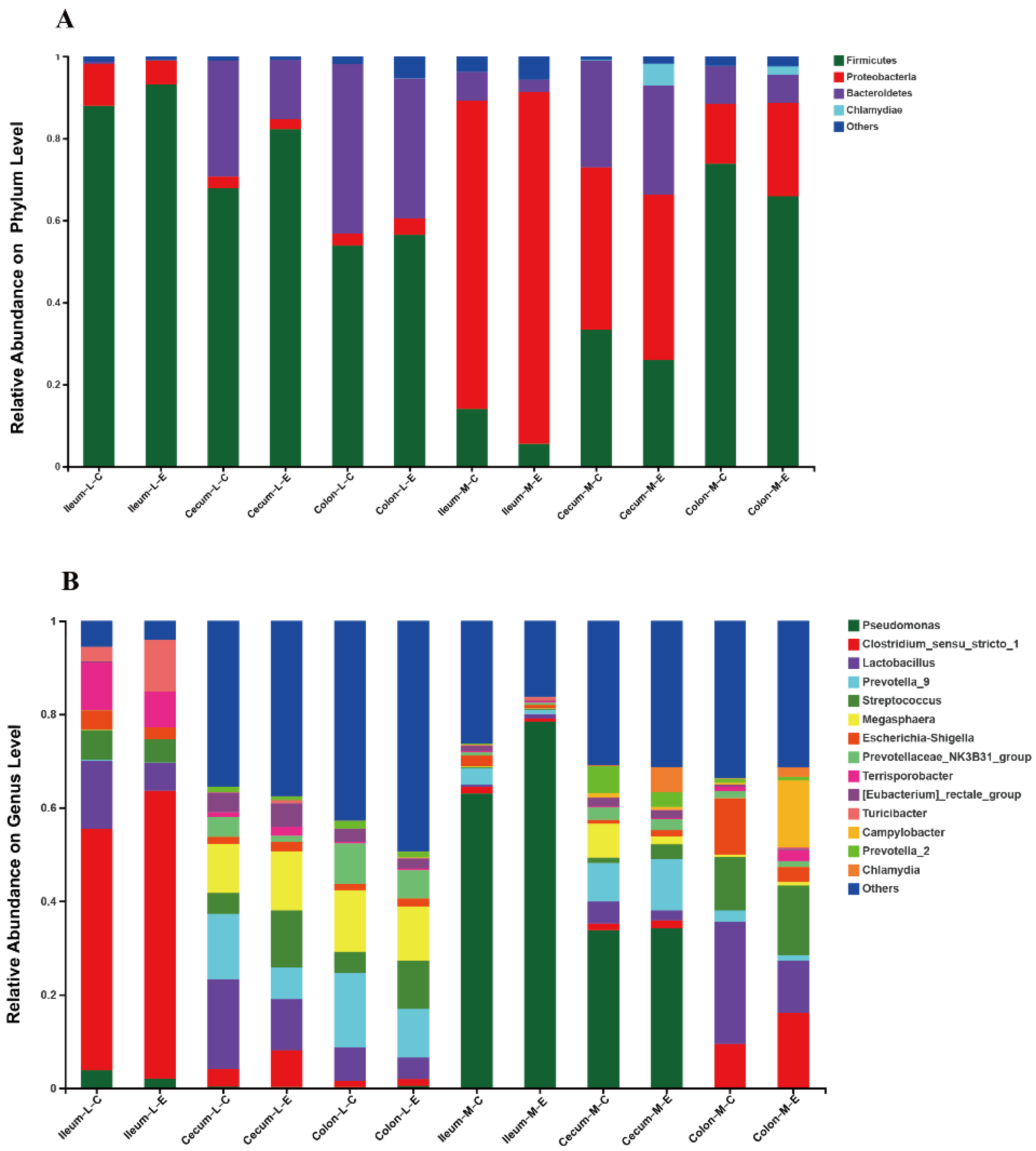
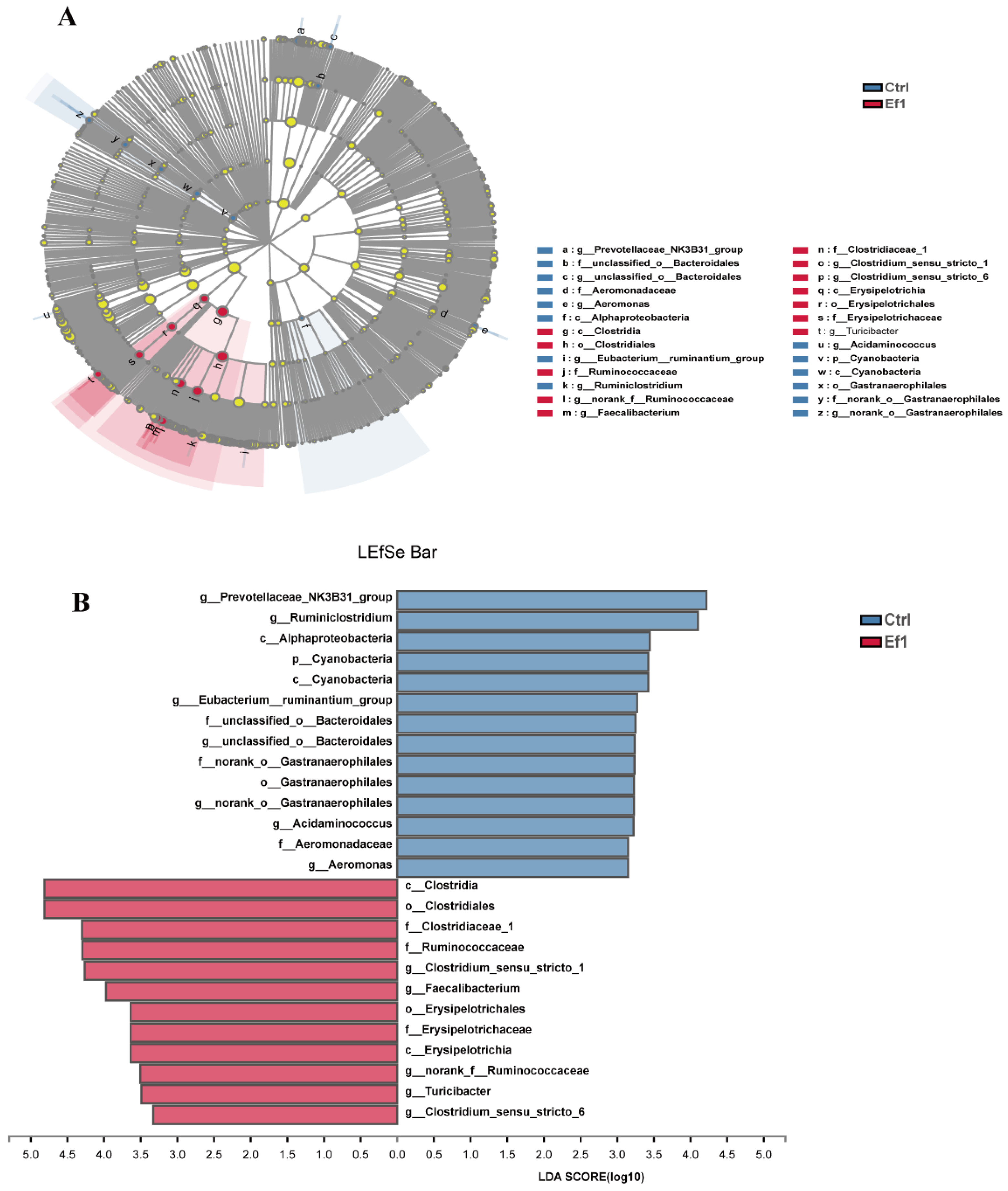
3.5. Effects of HDRsEf1 on Microbiota Composition in Different Niches
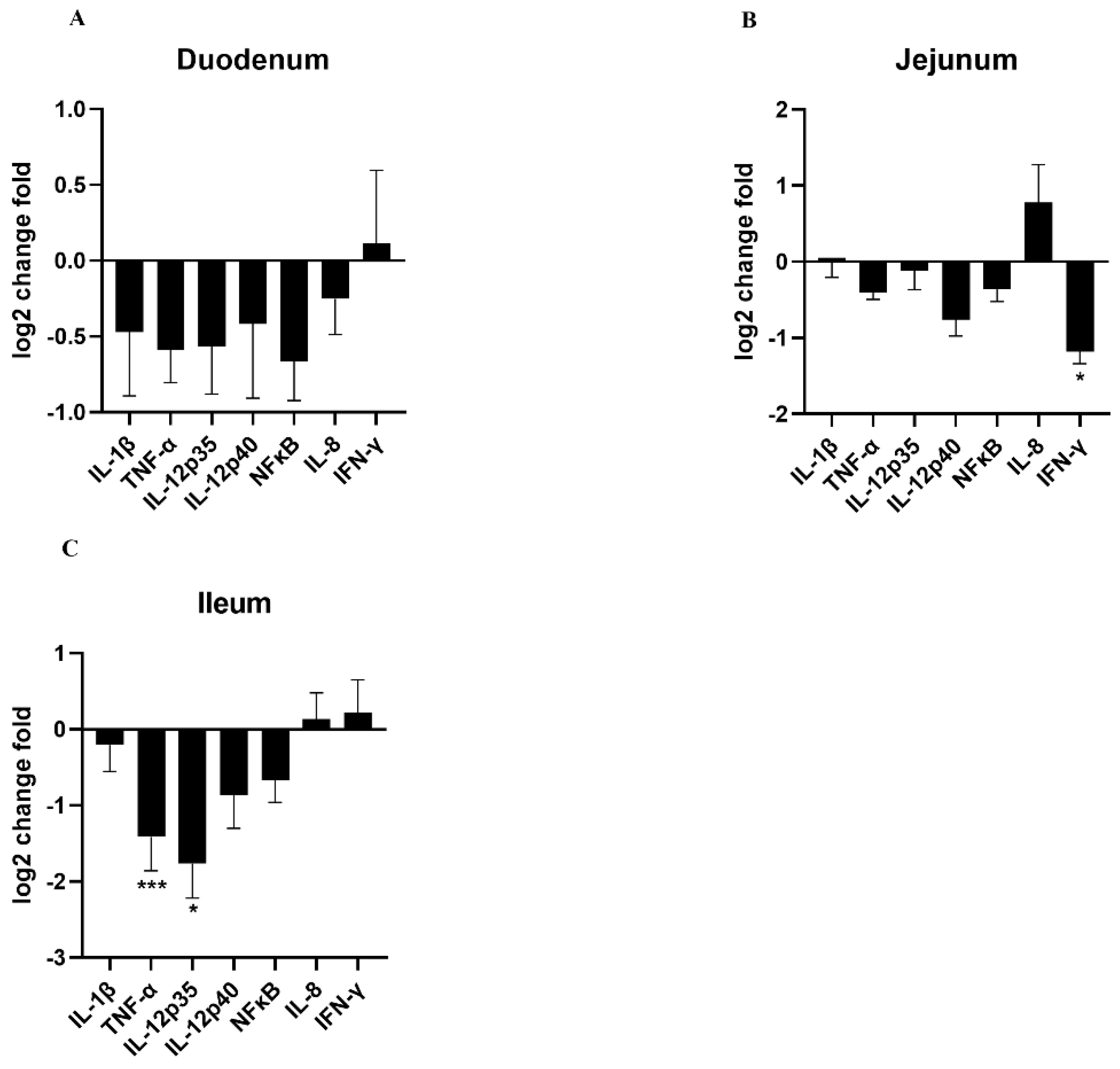
3.6. Effects of HDRsEf1 on Intestinal Inflammation
3.7. Correlation Analysis of Gut Microbiota Alterations and Inflammatory Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lalles, J.P.; Bosi, P.; Smidt, H.; Stokes, C.R. Nutritional management of gut health in pigs around weaning. Proc. Nutr. Soc. 2007, 66, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Casewell, M.; Friis, C.; Marco, E.; McMullin, P.; Phillips, I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 2003, 52, 159–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, M.; Smidt, H.; Mengheri, E.; Miller, B. Probiotics—Do they have a role in the pig industry? Animal 2011, 5, 462–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Shi, D.; Xiao, Y.; Bi, D.; Xiong, Y.; Wang, X.; Gao, X.; Li, Z.; Zhou, Z.; Liu, M.; Xu, Q.; et al. A Beneficial Enterococcus Faecium Strain’s Screening and Application. China Patent ZL201,110,452,087.2, 11 November 2013. [Google Scholar]
- Klingspor, S.; Martens, H.; Caushi, D.; Twardziok, S.; Lodemann, U. Characterization of the effects of Enterococcus faecium on intestinal epithelial transport properties in piglets. J. Anim. Sci. 2013, 91, 1707–1718. [Google Scholar] [CrossRef]
- Lodemann, U.; Hübener, K.; Jansen, N.; Martens, H. Effects of Enterococcus faecium NCIMB 10415 as probiotic supplement on intestinal transport and barrier function of piglets. Arch. Anim. Nutr. 2006, 60, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Bednorz, C.; Guenther, S.; Oelgeschlager, K.; Kinnemann, B.; Pieper, R.; Hartmann, S.; Tedin, K.; Semmler, T.; Neumann, K.; Schierack, P. Feeding the Probiotic Enterococcus faecium Strain NCIMB 10,415 to Piglets Specifically Reduces the Number of Escherichia coli Pathotypes That Adhere to the Gut Mucosa. Appl. Environ. Microbiol. 2013, 79, 7896–7904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollmann, M.; Nordhoff, M.; Pospischil, A.; Tedin, K.; Wieler, L.H. Effects of a Probiotic Strain of Enterococcus faecium on the Rate of Natural Chlamydia Infection in Swine. Infect. Immun. 2005, 73, 4346–4353. [Google Scholar] [CrossRef] [Green Version]
- Scharek, L.; Guth, J.; Reiter, K.; Weyrauch, K.D.; Taras, D.; Schwerk, P.; Schierack, P.; Schmidt, M.F.G.; Wieler, L.H.; Tedin, K. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet. Immunol. Immunopathol. 2005, 105, 151–161. [Google Scholar] [CrossRef]
- Capcarova, M.; Weiss, J.; Hrncar, C.; Kolesarova, A.; Pal, G. ORIGINAL ARTICLE: Effect of Lactobacillus fermentum and Enterococcus faecium strains on internal milieu, antioxidant status and body weight of broiler chickens. J. Anim. Physiol. Anim. Nutr. 2010, 94, e215–e224. [Google Scholar] [CrossRef]
- Fouhse, J.M.; Gao, J.; Vasanthan, T.; Izydorczyk, M.; Beattie, A.D.; Zijlstra, R.T. Whole-Grain Fiber Composition Influences Site of Nutrient Digestion, Standardized Ileal Digestibility of Amino Acids, and Whole-Body Energy Utilization in Grower Pigs. J. Nutr. 2017, 147, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.T.; Fukata, M.; Arditi, M. TLR Signaling in the Gut in Health and Disease. J. Immunol. 2005, 174, 4453–4460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeissig, S.; Blumberg, R.S. Life at the beginning: Perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 2014, 15, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.; McCracken, V.; Aminov, R.; Simpson, J.M.; Mackie, R.; Verstegen, M.; Gaskins, H.R. Gut microbiology and growth-promoting antibiotics in swine. Nutr. Abstr. Reviews. Ser. B Livest. Feeds Feed 2000, 70, 101–108. [Google Scholar]
- Dibner, J.J.; Richards, J.D. Antibiotic Growth Promoters in Agriculture: History and Mode of Action. Poult. Sci. 2005, 84, 634–643. [Google Scholar] [CrossRef]
- Li, P.; Niu, Q.; Wei, Q.; Zhang, Y.; Ma, X.; Kim, S.W.; Lin, M.; Huang, R. Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus Faecalis as alternatives to antibiotics. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Wang, T.; Teng, K.; Liu, Y.; Shi, W.; Zhang, J.; Dong, E.; Zhang, X.; Tao, Y.; Zhong, J. Lactobacillus plantarum PFM 105 Promotes Intestinal Development Through Modulation of Gut Microbiota in Weaning Piglets. Front. Microbiol. 2019, 10, 90. [Google Scholar] [CrossRef]
- Yang, S.C.; Lin, C.H.; Sung, C.T.; Fang, J.Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Sakaguchi, E. Transport of bacteria across and along the large intestinal lumen of guinea pigs. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2006, 176, 173–178. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, W.; Lee, Y.K.; Xie, J.; Zhang, H. Spatial Heterogeneity and Co-occurrence of Mucosal and Luminal Microbiome across Swine Intestinal Tract. Front. Microbiol. 2018, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bruni, L.; Jaramillo-Torres, A.; Gajardo, K.; Kortner, T.M.; Krogdahl, Å. Differential response of digesta- and mucosa-associated intestinal microbiota to dietary insect meal during the seawater phase of Atlantic salmon. Anim. Microbiome 2021, 3, 8. [Google Scholar] [CrossRef]
- Kelly, J.; Daly, K.; Moran, A.W.; Ryan, S.; Bravo, D.; Shirazi-Beechey, S.P. Composition and diversity of mucosa-associated microbiota along the entire length of the pig gastrointestinal tract; dietary influences. Environ. Microbiol. 2017, 19, 1425–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, F.; Clark, J.B. Early weaning stress impairs development of mucosal barrier function in the porcine intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G352–G363. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, Q.; Zhou, M.; Luo, Z.; Lv, L.; Pei, J.; Wang, C.; Chai, B.; Sui, B.; Huang, F.; et al. Composition of the murine gut microbiome impacts humoral immunity induced by rabies vaccines. Clin. Transl. Med. 2020, 10, e161. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, J.; Feng, P.; Zhu, W.; Mao, S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci. Rep. 2016, 6, 20329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Weber, C.F. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0-10. 2013. Available online: http://vegan.r-forge.r-project.org/ (accessed on 11 December 2019).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2015. Available online: http://th.archive.ubuntu.com/cran/web/packages/pheatmap/ (accessed on 28 January 2021).
- Pieper, R.; Bindelle, J.; Rossnagel, B.; Van Kessel, A.; Leterme, P. Effect of Carbohydrate Composition in Barley and Oat Cultivars on Microbial Ecophysiology and Proliferation of Salmonella enterica in an In Vitro Model of the Porcine Gastrointestinal Tract. Appl. Environ. Microbiol. 2009, 75, 7006–7016. [Google Scholar] [CrossRef] [Green Version]
- Mac, K.; Wichmann-Schauer, H.; Peters, J.; Ellerbroek, L. Species identification and detection of vancomycin resistance genes in enterococci of animal origin by multiplex PCR. Int. J. Food Microbiol. 2003, 88, 305–309. [Google Scholar] [CrossRef]
- Valeriano, V.D.; Balolong, M.P.; Kang, D.K. Probiotic roles of Lactobacillus sp. in swine: Insights from gut microbiota. J. Appl. Microbiol. 2017, 122, 554–567. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, A.M.; Yu, M.; Darby, T.M.; Vaccaro, C.; Li, J.Y.; Owens, J.A.; Hsu, E.; Adams, J.; Weitzmann, M.N.; Jones, R.M.; et al. The Microbial Metabolite Butyrate Stimulates Bone Formation via T Regulatory Cell-Mediated Regulation of WNT10B Expression. Immunity 2018, 49, 1116–1131. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a Cell Wall-Degrading Bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2018, 69, 2975–2984. [Google Scholar] [CrossRef] [Green Version]
- Gaskins, H.R.; Collier, C.T.; Anderson, D.B. Antibiotics as growth promotants: Mode of action. Anim. Biotechnol. 2002, 13, 29–42. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, X.; Ran, D.; Xiao, Y.; Wang, X.; Bi, D.; Shi, D. Enterococcus faecium HDRsEf1 Protects the Intestinal Epithelium and Attenuates ETEC-Induced IL-8 Secretion in Enterocytes. Mediat. Inflamm. 2016, 2016, 7474306. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Wang, R.; Hu, L.; Zhou, Q.; Liu, Y.; Yang, M.; Fang, Z.; Lin, Y.; Xu, S.; Feng, B. Enterococcus faecium NCIMB 10415 administration improves the intestinal health and immunity in neonatal piglets infected by enterotoxigenic Escherichia coli K88. J. Anim. Sci. Biotechnol. 2019, 10, 72. [Google Scholar] [CrossRef]
- Kaper, J.B. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef] [Green Version]
- Fairbrother, J.M.; Nadeau, E.; Gyles, C.L. Escherichia coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Wei, A.; Wang, H.; Zhang, X. Prevalence and characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front. Microbiol. 2015, 6, 1103. [Google Scholar] [CrossRef]
- Pruthvishree, B.S.; Kumar, O.R.V.; Sinha, D.K.; Malik, Y.P.S.; Dubal, Z.B.; Desingu, P.A.; Shivakumar, M.; Krishnaswamy, N.; Singh, B.R. Spatial molecular epidemiology of carbapenem-resistant and New Delhi metallo beta-lactamase (blaNDM)-producing Escherichia coli in the piglets of organized farms in India. J. Appl. Microbiol. 2017, 122, 1537–1546. [Google Scholar] [CrossRef]
- Devriendt, B.; Stuyven, E.; Verdonck, F.; Goddeeris, B.M.; Cox, E. Enterotoxigenic Escherichia coli (K88) induce proinflammatory responses in porcine intestinal epithelial cells. Dev. Comp. Immunol. 2010, 34, 1175–1182. [Google Scholar] [CrossRef]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 2009, 1788, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2018, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mccracken, B.A.; Spurlock, M.E.; Roos, M.A.; Zuckermann, F.A.; Gaskins, H.R. Weaning anorexia may contribute to local inflammation in the piglet small intestine. J. Nutr. 1999, 129, 613–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]

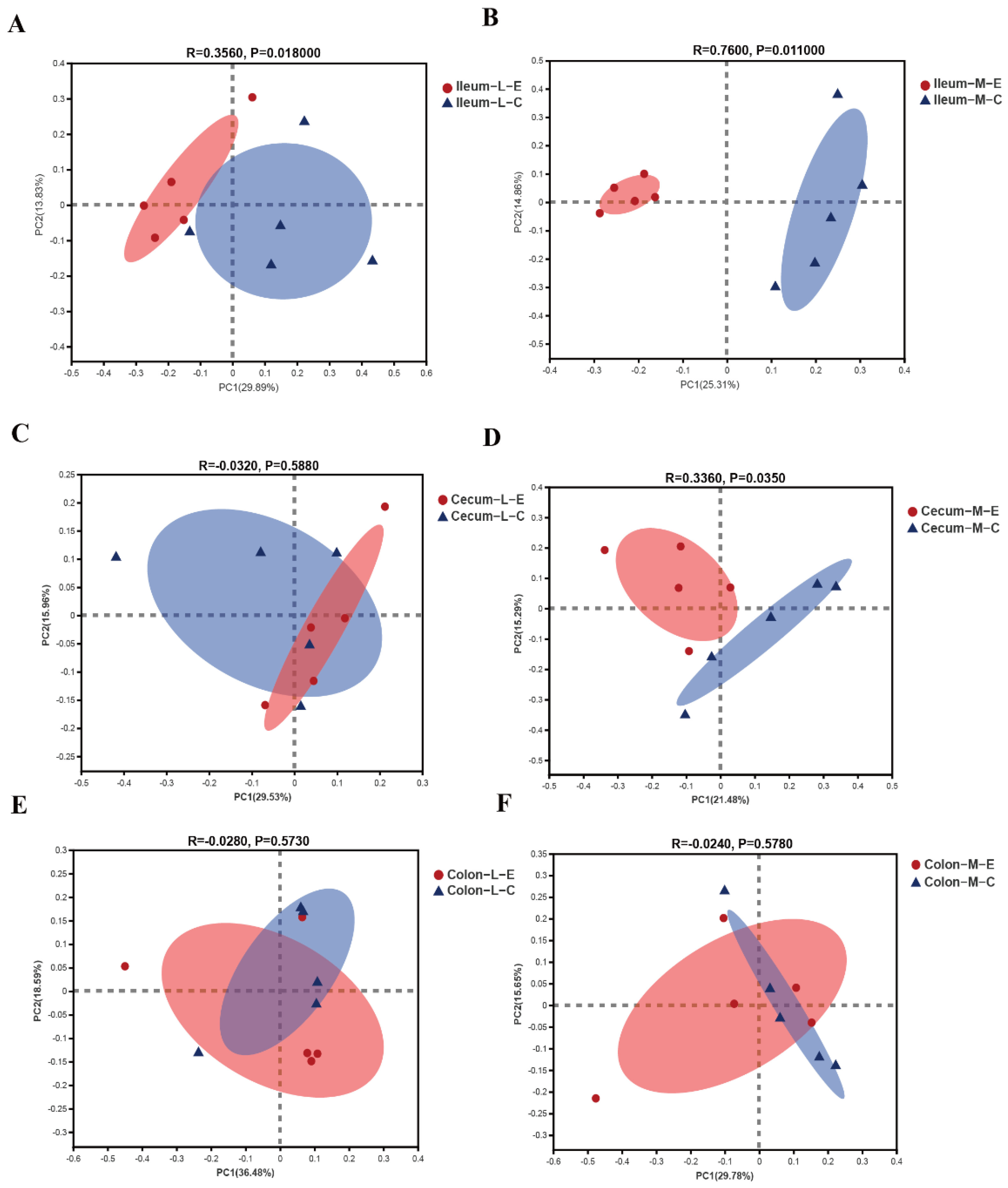



| Variables | Unweighted (R) | p-Value | Weighted (R) | p-Value |
|---|---|---|---|---|
| C VS E | ||||
| Ileal Lumen | 0.3560 | 0.018 | 0,024 | 0.348 |
| Ileal Mucosa | 0.7600 | 0.011 | 0.4600 | 0.014 |
| Cecal Lumen | −0.0320 | 0.588 | 0.3160 | 0.068 |
| Cecal Mucosa | 0.336 | 0.035 | −0.052 | 0.430 |
| Colonic Lumen | −0.028 | 0.573 | 0.0760 | 0.691 |
| Colonic Mucosa | −0.024 | 0.578 | 0.000 | 0.793 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Luo, J.; Yang, S.; Xiao, Q.; Wang, X.; Zhou, Z.; Xiao, Y.; Shi, D. Different Responses of Microbiota across Intestinal Tract to Enterococcus faecium HDRsEf1 and Their Correlation with Inflammation in Weaned Piglets. Microorganisms 2021, 9, 1767. https://doi.org/10.3390/microorganisms9081767
Zhou J, Luo J, Yang S, Xiao Q, Wang X, Zhou Z, Xiao Y, Shi D. Different Responses of Microbiota across Intestinal Tract to Enterococcus faecium HDRsEf1 and Their Correlation with Inflammation in Weaned Piglets. Microorganisms. 2021; 9(8):1767. https://doi.org/10.3390/microorganisms9081767
Chicago/Turabian StyleZhou, Jin, Ji Luo, Shumin Yang, Qiling Xiao, Xiliang Wang, Zutao Zhou, Yuncai Xiao, and Deshi Shi. 2021. "Different Responses of Microbiota across Intestinal Tract to Enterococcus faecium HDRsEf1 and Their Correlation with Inflammation in Weaned Piglets" Microorganisms 9, no. 8: 1767. https://doi.org/10.3390/microorganisms9081767









