Drug Resistance Determinants in Clinical Isolates of Enterococcus faecalis in Bangladesh: Identification of Oxazolidinone Resistance Gene optrA in ST59 and ST902 Lineages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Detection of Drug Resistance Genes
2.4. Genetic Analysis of aac(6′)-Ie-aph(2″)-Ia
2.5. Genetic Determinants of Oxazolidinone and Daptomycin Resistance
2.6. Multilocus Sequence Typing (MLST)
2.7. GenBank Accession Numbers
3. Results
3.1. Prevalence of Antimicrobial Resistance and Resistance Determinants
3.2. Genetic Analysis of aac(6′)-Ie-aph(2″)-Ia
3.3. ST of Isolates with Different Characteristics
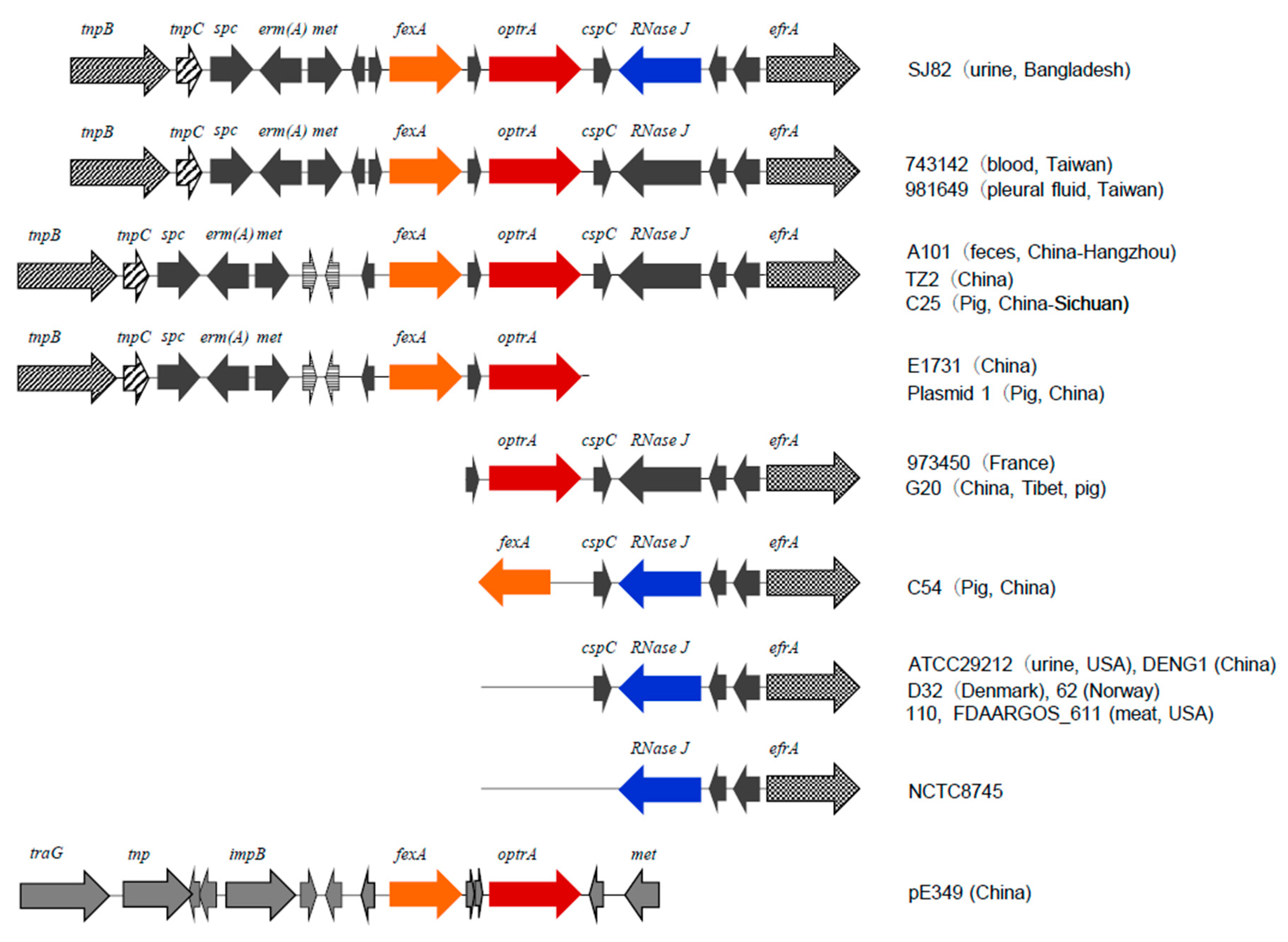
3.4. Genetic Background of fexA and optrA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beganovic, M.; Luther, M.K.; Rice, L.B.; Arias, C.A.; Rybak, M.J.; LaPlante, K.L. A Review of Combination Antimicrobial Therapy for Enterococcus faecalis Bloodstream Infections and Infective Endocarditis. Clin. Infect. Dis. 2018, 67, 303–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 123–184. [Google Scholar]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updat. 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Hines, K.M.; Waalkes, A.; Penewit, K.; Holmes, E.A.; Salipante, S.J.; Werth, B.J.; Xu, L. Characterization of the Mechanisms of Daptomycin Resistance among Gram-Positive Bacterial Pathogens by Multidimensional Lipidomics. mSphere 2017, 2, e00492-17. [Google Scholar] [CrossRef] [Green Version]
- Bi, R.; Qin, T.; Fan, W.; Ma, P.; Gu, B. The emerging problem of linezolid-resistant enterococci. J. Glob. Antimicrob. Resist. 2018, 13, 11–19. [Google Scholar] [CrossRef]
- Baddour, L.M.; Wilson, W.R.; Bayer, A.S.; Fowler, V.G., Jr.; Tleyjeh, I.M.; Rybak, M.J.; Barsic, B.; Lockhart, P.B.; Gewitz, M.H.; Levison, M.E.; et al. American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council. Infective endocarditis in adults: Diagnosis, antimicrobial therapy, and management of complications: A scientific statement for healthcare professionals from the American Heart Association. Circulation 2015, 132, 1435–1486. [Google Scholar]
- Fiedler, S.; Bender, J.K.; Klare, I.; Halbedel, S.; Grohmann, E.; Szewzyk, U.; Werner, G. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 2016, 71, 871–881. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.; Rabbi, M.B.; Sultana, S. Antibiotic resistance in Bangladesh: A systematic review. Int. J. Infect. Dis. 2019, 80, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Kawaguchiya, M.; Ghosh, S.; Paul, S.K.; Urushibara, N.; Mahmud, C.; Nahar, K.; Hossain, M.A.; Kobayashi, N. Drug resistance and molecular epidemiology of aerobic bacteria isolated from puerperal infections in Bangladesh. Microb. Drug Resist. 2015, 21, 297–306. [Google Scholar] [CrossRef]
- Haque, R.; Akter, M.L.; Salam, M.A. Prevalence and susceptibility of uropathogens: A recent report from a teaching hospital in Bangladesh. BMC Res. Notes 2015, 8, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhter, J.; Ahmed, S.; Saleh, A.A.; Anwar, S. Antimicrobial resistance and in vitro biofilm-forming ability of Enterococci spp. isolated from urinary tract infection in a tertiary care hospital in Dhaka. Bangladesh Med. Res. Counc. Bull. 2014, 40, 6–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, N.; Alam, M.; Nishimoto, Y.; Urasawa, S.; Uehara, N.; Watanabe, N. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 2001, 126, 197–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahbub Alam, M.; Kobayashi, N.; Ishino, M.; Sumi, A.; Kobayashi, K.; Uehara, N.; Watanabe, N. Detection of a novel aph(2") allele (aph [2"]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad 9) in clinical isolates of enterococci. Microb. Drug Resist. 2005, 11, 239–247. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 24th Informational Supplement (M100-S25); Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0, 2020. Available online: http://www.eucast.org (accessed on 1 July 2020).
- Tamang, M.D.; Moon, D.C.; Kim, S.R.; Kang, H.Y.; Lee, K.; Nam, H.M.; Jang, G.C.; Lee, H.S.; Jung, S.C.; Lim, S.K. Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet. Microbiol. 2017, 201, 252–256. [Google Scholar] [CrossRef]
- Isogai, N.; Urushibara, N.; Kawaguchiya, M.; Ghosh, S.; Suzaki, K.; Watanabe, N.; Quiñones, D.; Kobayashi, N. Characterization of Enterococcus faecium with macrolide resistance and reduced susceptibility to quinupristin/dalfopristin in a Japanese hospital: Detection of extensive diversity in erm(B)-regulator regions. Microb. Drug Resist. 2013, 19, 298–307. [Google Scholar] [CrossRef]
- Depardieu, F.; Perichon, B.; Courvalin, P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 2004, 42, 5857–5860. [Google Scholar] [CrossRef] [Green Version]
- Nishimoto, Y.; Kobayashi, N.; Alam, M.M.; Ishino, M.; Uehara, N.; Watanabe, N. Analysis of the prevalence of tetracycline resistance genes in clinical isolates of Enterococcus faecalis and Enterococcus faecium in a Japanese hospital. Microb. Drug Resist. 2005, 11, 146–153. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [Green Version]
- Oyamada, Y.; Ito, H.; Inoue, M.; Yamagishi, J.I. Topoisomerase mutations and efflux are associated with fluoroquinolone resistance in Enterococcus faecalis. J. Med. Microbiol. 2006, 55, 1395–1401. [Google Scholar] [CrossRef]
- Watanabe, S.; Kobayashi, N.; Quiñones, D.; Nagashima, S.; Uehara, N.; Watanabe, N. Genetic diversity of enterococci harboring the high-level gentamicin resistance gene aac(6′)-Ie-aph(2″)-Ia or aph(2″)-Ie in a Japanese hospital. Microb. Drug Resist. 2009, 15, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgeois-Nicolaos, N.; Massias, L.; Couson, B.; Butel, M.J.; Andremont, A.; Doucet-Populaire, F. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J. Infect. Dis. 2007, 195, 1480–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, L.; Kiratisin, P.; Mendes, R.E.; Panesso, D.; Singh, K.V.; Arias, C.A. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 2012, 56, 3917–3922. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Garbajosa, P.; Bonten, M.J.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Cantón, R.; Baquero, F.; Murray, B.E.; et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Schwarz, S.; Chi, D.; Wang, Z.; Zhang, R.; Wang, Y. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. 2019, 25, 630.e1–630.e6. [Google Scholar] [CrossRef]
- Deshpande, L.M.; Castanheira, M.; Flamm, R.K.; Mendes, R.E. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018, 73, 2314–2322. [Google Scholar] [CrossRef] [Green Version]
- Mendes, R.E.; Sader, H.S.; Castanheira, M.; Flamm, R.K. Distribution of main Gram-positive pathogens causing bloodstream infections in United States and European hospitals during the SENTRY Antimicrobial Surveillance Program (2010–2016): Concomitant analysis of oritavancin in vitro activity. J. Chemother. 2018, 30, 280–289. [Google Scholar] [CrossRef] [Green Version]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. Antimicrob. Resist. Infect. Control. 2019, 8, 55. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Mendes, R.E.; Streit, J.M.; Hogan, P.A.; Flamm, R.K. ZAAPS Program results for 2015: An activity and spectrum analysis of linezolid using clinical isolates from medical centres in 32 countries. J. Antimicrob. Chemother. 2017, 72, 3093–3099. [Google Scholar] [CrossRef]
- Das, A.; Banerjee, T.; Anupurba, S. Susceptibility of Nitrofurantoin and Fosfomycin against Outpatient Urinary Isolates of Multidrug-Resistant Enterococci over a Period of 10 Years from India. Microb. Drug Resist. 2019. [Google Scholar] [CrossRef] [PubMed]
- Osuka, H.; Nakajima, J.; Oishi, T.; Funayama, Y.; Ebihara, T.; Ishikawa, H.; Saito, K.; Koganemaru, H.; Hitomi, S. High-level aminoglycoside resistance in Enterococcus faecalis and Enterococcus faecium causing invasive infection: Twelve-year surveillance in the Minami Ibaraki Area. J. Infect. Chemother. 2016, 22, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Coombs, G.W.; Daley, D.A.; Mowlaboccus, S.; Lee, Y.T.; Pang, S. Australian Group on Antimicrobial Resistance. Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2018. Commun. Dis. Intell. 2020, 44. [Google Scholar] [CrossRef]
- Haghi, F.; Lohrasbi, V.; Zeighami, H. High incidence of virulence determinants, aminoglycoside and vancomycin resistance in enterococci isolated from hospitalized patients in Northwest Iran. BMC Infect. Dis. 2019, 19, 744. [Google Scholar] [CrossRef] [Green Version]
- Lyon, B.R.; Gillespie, M.T.; Skurray, R.A. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 1987, 133, 3031–3038. [Google Scholar] [CrossRef] [Green Version]
- Maki, H.; Murakami, K. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997, 179, 6944–6948. [Google Scholar] [CrossRef] [Green Version]
- Leelaporn, A.; Yodkamol, K.; Waywa, D.; Pattanachaiwit, S. A novel structure of Tn4001-truncated element, type V, in clinical enterococcal isolates and multiplex PCR for detecting aminoglycoside resistance genes. Int. J. Antimicrob. Agents 2008, 31, 250–254. [Google Scholar] [CrossRef]
- Zhang, J.M.; Wang, Q.; Han, T.Y.; Liu, J.H.; Hu, X.X.; Qiao, F.; Yang, X.Y.; Li, C.R.; You, X.F. Structure analysis of transposons carrying the aac(6′)-aph(2″) gene in Enterococcus faecalis isolated in Beijing, China, and comparison of their transfer efficiency. Int. J. Antimicrob. Agents 2018, 52, 799–804. [Google Scholar] [CrossRef]
- Sharkey, L.K.R.; O’Neill, A.J. Antibiotic Resistance ABC-F Proteins: Bringing Target Protection into the Limelight. ACS Infect. Dis. 2018, 4, 239–246. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Yin, Y.; Li, S.; Zhang, Y.; Wang, Q.; Wang, H. Molecular characteristics of oxazolidinone resistance in enterococci from a multicenter study in China. BMC Microbiol. 2019, 19, 162. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, G.; Li, J.; Chen, L.; Liu, H.; Bi, W.; Lu, H.; Zhou, T. A high incidence and coexistence of multiresistance genes cfr and optrA among linezolid-resistant enterococci isolated from a teaching hospital in Wenzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wang, Y.; Schwarz, S.; Lv, H.; Li, Y.; Liao, K.; Yu, S.; Zhao, K.; Gu, D.; Wang, X.; et al. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010–2014. Clin. Microbiol. Infect. 2015, 21, 1095.e1–1095.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, L.; Wang, Y.; Lv, Y.; Wang, S.; Song, Y.; Li, Y.; Liu, J.; Xue, F.; Yang, W.; Zhang, J. Nationwide Surveillance of Novel Oxazolidinone Resistance Gene optrA in Enterococcus Isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 2016, 60, 7490–7493. [Google Scholar] [PubMed] [Green Version]
- Mendes, R.E.; Deshpande, L.; Streit, J.M.; Sader, H.S.; Castanheira, M.; Hogan, P.A.; Flamm, R.K. ZAAPS programme results for 2016: An activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J. Antimicrob. Chemother. 2018, 73, 1880–1887. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Jeong, Y.S.; Chang, J.; Sung, H.; Kim, M.N. Emergence of optrA-Mediated Linezolid-Nonsusceptible Enterococcus faecalis in a Tertiary Care Hospital. Ann. Lab. Med. 2020, 40, 321–325. [Google Scholar] [CrossRef] [Green Version]
- Bender, J.K.; Fleige, C.; Lange, D.; Klare, I.; Werner, G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 2018, 52, 819–827. [Google Scholar] [CrossRef]
- Chen, M.; Pan, H.; Lou, Y.; Wu, Z.; Zhang, J.; Huang, Y.; Yu, W.; Qiu, Y. Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect. Drug Resist. 2018, 11, 2397–2409. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Gao, S.; Xu, H.; Zhang, Z.; Chen, F.; Shen, H.; Zhang, C. Distribution of the optrA gene in Enterococcus isolates at a tertiary care hospital in China. J. Glob. Antimicrob. Resist. 2019, 17, 180–186. [Google Scholar] [CrossRef]
- Almeida, L.M.; Lebreton, F.; Gaca, A.; Bispo, P.M.; Saavedra, J.T.; Calumby, R.N.; Grillo, L.M.; Nascimento, T.G.; Filsner, P.H.; Moreno, A.M.; et al. Transferable Resistance Gene optrA in Enterococcus faecalis from Swine in Brazil. Antimicrob. Agents Chemother. 2020, 64, e00142-20. [Google Scholar] [CrossRef]
- Elghaieb, H.; Tedim, A.P.; Abbassi, M.S.; Novais, C.; Duarte, B.; Hassen, A.; Peixe, L.; Freitas, A.R. From farm to fork: Identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals and retail meat. J. Antimicrob. Chemother. 2020, 75, 30–35. [Google Scholar] [CrossRef]
- Li, D.; Li, X.Y.; Schwarz, S.; Yang, M.; Zhang, S.M.; Hao, W.; Du, X.D. Tn6674 Is a Novel Enterococcal optrA -Carrying Multiresistance Transposon of the Tn554 Family. Antimicrob. Agents Chemother. 2019, 63, e00809-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Antimicrobials 1/Resistance Determinants 2 | Number of Resistant Isolates/Isolates with Resistant Determinant (%) (n = 210) | |
|---|---|---|
| Antimicrobial agents | ||
| AMP | 0 (0) | |
| IPM | 0 (0) | |
| GEN-HLR | 24 (11.4) | |
| VAN | 0 (0) | |
| TEC | 0 (0) | |
| ERY | 179 (85.2) | |
| TET | 125 (59.5) | |
| MIN | 17 (8.1) | |
| NIT | 22 (10.5) | |
| FOF | 0 (0) | |
| LVX | 96 (45.7) | |
| LZD | 5 4 (2.4) | |
| Resistance gene/determinant | ||
| (Aminoglycoside) | aac(6′)-Ie-aph(2″)-Ia | 46 (21.9) |
| aph(3′)-IIIa | 73 (34.8) | |
| ant(6)-Ia | 18 (8.6) | |
| ant(9)-Ia | 8 (3.8) | |
| (Macrolide) | erm(A) | 1 (0.5) |
| erm(B) | 204 (97.1) | |
| (Tetracycline) | tet(L) | 80 (38.1) |
| tet(M) | 128 (61.0) | |
| (Oxazolidinone) | fexA-optrA | 5 (2.4) |
| (QRDR 3 mutation) | GyrA: S 84 I and ParC: S 82 I | 93 (44.3) |
| GyrA: S 84 I | 3 (1.4) | |
| Isolate ID | Age/Sex of Patient | Antimicrobial Resistance Pattern 1 | Drug Resistance Genes 2 | MIC (μg/mL) of GEN | IS256 Flanking Pattern (aac-(6′)-Ie-aph(2″)-Ia) | MIC (μg/mL) of LVX | QRDR Mutation 3 | Sequence Type 4 (MLST) | Clonal Complex (CC), ST Variant | |
|---|---|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | |||||||||
| SJ5 | 70/M | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | A | 64 | S 84 I | S 82 I | ST28 | CC28 |
| SJ42 | 3/F | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | A | 64 | S 84 I | S 82 I | ST28 | CC28 |
| SJ94 | 45/M | ERY, GEN-HLR, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, ant(6)-Ia, erm(B), tet(M) | >1024 | A | 32 | S 84 I | S 82 I | ST28 | CC28 |
| SJ32 | 30/F | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(M) | >1024 | A | 128 | S 84 I | S 82 I | ST28 | CC28 |
| SJ238 | 38/M | ERY, GEN-HLR, LVX, TET, KAN | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(L), tet(M) | >1024 | A | 32 | S 84 I | S 82 I | ST946 | CC116 |
| SJ125 | 40/M | ERY, GEN-HLR, LVX, TET, MIN, KAN | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, ant(9)-Ia, erm(A), erm(B), tet(L), tet(M) | >1024 | A | 32 | S 84 I | S 82 I | ST6 | CC6 |
| SJ204 | 3.5/F | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | A | 32 | S 84 I | S 82 I | ST6 | CC6 |
| SJ40 | 5/F | ERY, GEN-HLR, LVX, TET, MIN, NIT | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | >1024 | A | 64 | S 84 I | S 82 I | ST6 | CC6 |
| SJ127 | 18/M | ERY, GEN-HLR, KAN, LVX, TET, MIN, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | A | 32 | S 84 I | S 82 I | ST6 | CC6 |
| SJ208 | 20/F | ERY, GEN-HLR, KAN, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | < 2 | NM | NM | ST363 | CC16 |
| SJ207 | 55/M | ERY, GEN-HLR, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(L), tet (M) | >1024 | B | 64 | S 84 I | S 82 I | ST28 | CC28 |
| SJ3 | 40/F | ERY, GEN-HLR, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | 128 | S 84 I | S 82 I | ST28 | CC28 |
| SJ10 | 2/M | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | 64 | S 84 I | S 82 I | ST6 | CC6 |
| SJ8 | 47/F | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | 128 | S 84 I | S 82 I | ST28 | CC28 |
| SJ11 | 22/F | ERY, GEN-HLR, KAN, LVX, TET, NIT | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | 64 | S 84 I | S 82 I | ST965 * | ST919 SLV |
| SJ13 | 2/M | ERY, GEN-HLR, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | B | 64 | S 84 I | S 82 I | ST966 * | CC28 |
| SJ38 | 5/F | ERY, GEN-HLR, KAN, LVX, TET, MIN | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(L), tet(M) | >1024 | C | 16 | S 84 I | S 82 I | ST28 | CC28 |
| SJ77 | 65/M | TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(L), tet(M) | 64 | C | < 2 | NM | NM | ST947 * | CC116 |
| SJ92 | 33/F | ERY, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(L), tet(M) | 4 | C | 16 | S 84 I | S 82 I | ST947 * | CC116 |
| SJ95 | 50/M | ERY, GEN-HLR, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(L), tet(M) | >1024 | C | 32 | S 84 I | S 82 I | ST947 * | CC116 |
| SJ96 | 32/F | ERY, GEN-HLR, KAN, LVX, TET, MIN | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, erm(B), tet(L), tet(M) | >1024 | C | 16 | S 84 I | S 82 I | ST947 * | CC116 |
| SJ128 | 28/F | ERY, TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | 128 | D | < 2 | NM | NM | ST16 | CC16 |
| SJ132 | 30/F | ERY, TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | 128 | D | < 2 | NM | NM | ST16 | CC16 |
| SJ134 | 30/M | ERY, TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | 64 | D | < 2 | NM | NM | ST16 | CC16 |
| SJ31 | 1/F | ERY, GEN-HLR, KAN, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | >1024 | D | 32 | S 84 I | NM | ST415 | CC941 |
| SJ81 | 40/M | ERY, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | 32 | D | < 2 | NM | NM | ST16 | CC16 |
| SJ126 | 30/F | ERY, LVX, TET | aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) | 128 | D | < 2 | NM | NM | ST818 | CC16 |
| SJ218 | 11/F | LVX, TET, MIN | ant(9)-Ia, erm(B), erm(B), tet(L), tet(M) | 8 | 16 | S 84 I | NM | ST945 * | CC28 | |
| SJ71 | 7/F | ERY, KAN, TET, MIN | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M) | 8 | < 2 | NM | NM | ST21 | CC21 | |
| SJ28 | 55/F | ERY, KAN, LVX, TET | aph(3′)-IIIa, ant(6)-Ia, erm(B), tet(M) | 16 | 64 | S 84 I | S 82 I | ST28 | CC28 | |
| SJ69 | 27/F | ERY, KAN, TET | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(M) | 8 | < 2 | NM | NM | ST506 | CC100 | |
| SJ80 | 50/M | ERY | erm(B) | 8 | < 2 | NM | NM | ST919 * | ST28 TLV | |
| SJ148 | 33/F | ERY | erm(B), tet(L) | 4 | < 2 | NM | NM | ST919 * | ST28 TLV | |
| SJ52 | 24/F | ERY, TET | erm(B), tet(L), tet(M) | 2 | < 2 | NM | NM | ST919 * | ST28 TLV | |
| SJ87 | 20/M | ERY, KAN, LVX, TET, NIT, LZD | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M), fexA-optrA | 8 | 8 | S 84 I | S 82 I | ST59 | CC59 | |
| SJ88 | 18/M | ERY, KAN, LVX, TET, MIN, NIT, LZD | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M), fexA-optrA | 4 | 8 | S 84 I | S 82 I | ST59 | CC59 | |
| SJ82 | 25/F | ERY, KAN, TET, MIN, NIT, LZD | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M), fexA-optrA | 8 | < 2 | NM | NM | ST902 | ST21 TLV | |
| SJ117 | 50/M | ERY, KAN, TET, MIN, NIT, LZD | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M), fexA-optrA | 4 | < 2 | NM | NM | ST902 | ST21 TLV | |
| SJ116 | 28/F | ERY, KAN, LVX, TET, MIN, NIT, LZD, DAP | aph(3′)-IIIa, ant(9)-Ia, erm(B), tet(L), tet(M), fexA-optrA | 8 | 8 | S 84 I | S 82 I | ST917 * | CC59 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, S.; Aung, M.S.; Paul, S.K.; Ahmed, S.; Haque, N.; Khan, E.R.; Barman, T.K.; Islam, A.; Abedin, S.; Sultana, C.; et al. Drug Resistance Determinants in Clinical Isolates of Enterococcus faecalis in Bangladesh: Identification of Oxazolidinone Resistance Gene optrA in ST59 and ST902 Lineages. Microorganisms 2020, 8, 1240. https://doi.org/10.3390/microorganisms8081240
Roy S, Aung MS, Paul SK, Ahmed S, Haque N, Khan ER, Barman TK, Islam A, Abedin S, Sultana C, et al. Drug Resistance Determinants in Clinical Isolates of Enterococcus faecalis in Bangladesh: Identification of Oxazolidinone Resistance Gene optrA in ST59 and ST902 Lineages. Microorganisms. 2020; 8(8):1240. https://doi.org/10.3390/microorganisms8081240
Chicago/Turabian StyleRoy, Sangjukta, Meiji Soe Aung, Shyamal Kumar Paul, Salma Ahmed, Nazia Haque, Emily Rahman Khan, Tridip Kanti Barman, Arup Islam, Sahida Abedin, Chand Sultana, and et al. 2020. "Drug Resistance Determinants in Clinical Isolates of Enterococcus faecalis in Bangladesh: Identification of Oxazolidinone Resistance Gene optrA in ST59 and ST902 Lineages" Microorganisms 8, no. 8: 1240. https://doi.org/10.3390/microorganisms8081240





