1. Introduction
Tuberculosis (TB), a disease caused by the bacterium
Mycobacterium tuberculosis (Mtb), still remains the top infectious disease killer worldwide, ranking above HIV/AIDS [
1]. The gold standard for TB detection and diagnosis has always been culture-based. This old fashioned gold standard technique may be effective but suffers many limitations, especially when it comes to the rapid diagnosis of TB disease [
2]. Such limitations have driven scientists to come up with modernized techniques for TB diagnosis, including microscopic observation drug susceptibility (MODS) and molecular diagnostic systems [
3]. Polymerase chain reaction (PCR) is a molecular diagnostic technology that is commonly used in the amplification of specific molecular targets. Biotechnological refinements of PCR led to the development of technologies that not only detect targets but also quantify them [
4]. Quantification systems like the real-time quantification PCR (qPCR) and digital PCR (dPCR) were recently developed to quantify nucleic acid targets within a sample. Both these two are extensively used in the diagnosis of a variety of pathogens, including tuberculosis with dPCR, which shows many advantages over qPCR, as it does not need a standard curve during quantification [
4,
5].
Droplet digital PCR (ddPCR) is a third generation of PCR that was recently developed to overcome the disadvantage of dPCR during sample portioning [
2]. The ddPCR method works by separating a sample into thousands to millions of droplets depending on the system, and then partitioning them to be read as either positive or negative depending on fluorescence amplitude that is exhibited. Using poison statistics, droplets that contain target sequences (positive) versus those that do not contain any targets (negative) are used to calculate the exact quantity of nucleic acid targets within a sample in copies/µL, without generating a standard curve [
4]. Many methods are used in quantifying and detecting various targets using both EvaGreen and TaqMan probe assays for multiplexing in ddPCR. However, few studies exist in the application of ddPCR to detect
Mtb [
2]. In this study, two
Mtb specific targets,
IS6110 and
IS1081, were used for the detection and quantification of
Mtb using ddPCR.
Since their characterization, insertion sequences (
IS) have long been used in the diagnosis of
Mtb. The most commonly used
IS in tuberculosis diagnosis has always been
IS6110. This sequence, however, is reported to be missing in some clinical samples, hence giving room for false-negative [
6] and false-positive results [
7]. To curb the ineffectiveness of this sequence in large parts of the world, including Southeast Asia (Vietnam) [
8], another insertion sequence
IS1081 was used as a subsidiary marker in TB detection studies [
9,
10,
11]. In our study, we develop a duplex single dye droplet digital PCR assay capable of detecting the two genes reliably in TB samples. This study also shows steps using TaqMan probe assays to perform a single dye (FAM) duplex ddPCR experiment capable of detecting and quantifying the two genes in a single channel. To the best of our knowledge, no other research has combined these two genes using a single dye based ddPCR assay in detecting
Mtb.
2. Materials and Methods
2.1. Bacterial Strains and Ethical Considerations
Two bacterial strains M. tuberculosis H37Ra and Mycobacterium bovis bacillus Calmette-Guérin (BCG) were used for optimization in this study. Both the two Beijing strains were cultured and stored in our BSL 2 laboratory using solid agar (Difco™ Middlebrook 7H10 Agar) and liquid broth (Difco™ Middlebrook 7H9 Broth) medium. The conditions for reagent preparation and storage, including supplementation with OADC, were followed according manufacturer’s instructions. Another set of bacteria was provided by CDC China. Thirty samples labeled PT11HB01 to PT11HB30 and M. tuberculosis H37Rv were provided for testing. All the samples from CDC China were handled in our BSL 3 laboratory. No ethical permission for the study was sought as the Mtb samples were anonymized and no patient information could be retrieved from them.
2.2. Sample Processing
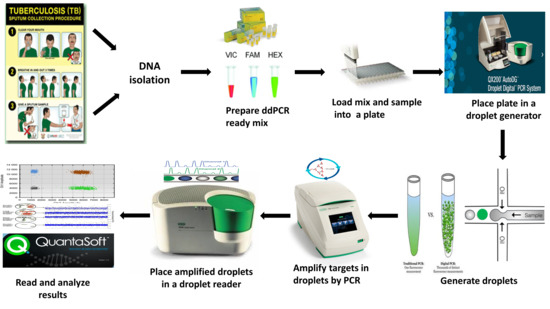
In order to study the effect of sample matrix on PCR reactions, three sputum mock samples were collected from healthy individuals and spiked with the two strains,
M. tuberculosis H37Ra and
Mycobacterium bovis BCG, to represent positive samples, and water used to represent controls. Spiked sputum samples and the controls were processed using the conventional N-acetyl-L-cysteine–sodium hydroxide (NALC–NaOH) method as described in the
Supplementary Methods for digestion and decontamination. Following decontamination, the bacterial suspension was used for various PCR tests (qPCR and ddPCR), as shown in
Figure 1.
2.3. DNA Extraction
To find a fast and efficient method for rapid DNA extraction involving minimal sample handling steps, a previously described method called thermocold lysis was modified [
3]. In this previous method, a sample was subjected to three cycles of heating at 95 °C for 20 min and freezing at −20 °C for 20 min. In our method, we sought to optimize the method using a commercially available, automated dry water bath (0.2 mL dry bath incubator model MK-20). The DNA extraction conditions were set for three cycles of heating at 95 °C for 20 min and cooling at −7 °C for 20 min. The minimum temperature was set at −7 °C, as the dry bath incubator could achieve a minimum temperature of up to −10 °C. After extraction, all the tubes were centrifuged at 10,000×
g at 4 °C for 10 min. The resultant supernatant was transferred to a new clean tube to be used in PCR experiments and/or stored at −20 °C until usage.
2.4. Assay Design and Optimization
Primers and probes specific to the two targets,
IS6110 and
IS1081, were adopted from Chakravorty et al. [
10]. The primer and probe sequences were used to generate amplicons shorter than 110 base pairs in length, as described in
Table S1. All the primers and probes used in the study were synthesized by the company Sangon Biotech, Shanghai, China. In order to optimize the conditions for
Mtb detection, different parameters including temperature gradient, primer, and probe concentration were tested to validate the assay performance. All these tests are described in detail in
Supplementary Methods.
2.5. Analytical Specificity and Sensitivity Tests
The specificity and exclusivity of the primers and probes for this assay were reported before on 30 different species of nontuberculous mycobacteria (NTM) and different common upper respiratory tract bacteria [
10]. In our test, we tested 10 different NTM (
Mycobacterium avium, Mycobacterium marinum, Mycobacterium shimoidei, Mycobacterium kansasii, Mycobacterium asiaticum, Mycobacterium scrofulaceum, Mycobacterium gordonae, Mycobacterium chelonae, Mycobacterium fortuitum, Mycobacterium phlei) and 5 respiratory tract bacteria (
Nocardia brasiliensis, Beijing corynebacterium, Legionella pneumophila, Bordetella pertussis, Pneumococcus) for specificity and exclusivity. All the primers used were tested for sensitivity online first by using the basic local alignment search tool (BLAST) from the national center for biotechnology information (NCBI) [
12]. After establishing the sensitivity, the assay capabilities to detect the two genes in singleplex and duplex assays were tested using laboratory-cultured
M. bovis BCG,
Mtb. H37Ra, and
Mtb. H37Rv.
2.6. Duplex Real-Time PCR Assay Composition
All qPCR experiments were performed in a CFX96 Touch™ real-time PCR detection system (CFX96, Bio-Rad Laboratories, Hercules, CA, USA). A 2× qPCR mixture with a final volume of 20 µL was used in all experiments. The composition of the mixture included 1.6 µL dNTP (2.5 mM for each, from Takara Biomedical Technology (Beijing) Co., Ltd, Beijing, China), 2 µL 10 × buffer (200 mM Tris-HCl pH 8.3, 200 mM KCl, 100 mM (NH4)2SO4, 20mM MgSO4 and 5% NONIDET P-40. SUBSTITUTE from AMRESCO Inc., Radnor, PA, USA), 0.5 µL Taq polymerase (activity unit 2.5 U/mL, expressed and purified in the laboratory), and 10.3 µL MilliQ water, 0.4 µL of 20 µM primer concentrations were added to the mixture and 1 µL of 10 µM probe was also added to the mixture. A set of two primers and probes (labeled at the 5′ end with FAM and 3′ end with BHQ 1) were used for detection. To reach a final volume of 20 µL, a sample volume of 2 µL was added. The qPCR reaction mixture was then amplified with an initial denaturation step at 95 °C for 5 s, followed by 40 cycles of denaturation at 95 °C for 5 s and an annealing (reading) step at 64 °C for 1 min.
2.7. Duplex Droplet Digital PCR Assay Composition
Both
IS6110 and
IS1081 copy numbers were quantified in samples using the automated QX200™ Droplet Digital™ PCR system (Bio-Rad, Hercules, CA, USA). Briefly, before preparing the reaction mix, the sample concentration was determined using a nanodrop machine (ND-2000, Thermo Fisher, Waltham, MA, USA). Concentrations ≤60 ng/µL were considered ideal for testing. If the concentrations were higher than this, they were diluted and redetermined until the final concentration was ≤60 ng/µL. The 22 µL ddPCR mixture was composed of 11 µL of 2 × ddPCR Supermix for Probes (No dUTP), 0.7 µL of 20 µM forward and reverse primers, 0.8 µL of 10 µM probes (both labeled at the 5′ end with FAM and 3′ end with BHQ 1), DNase/RNase free MilliQ water, and 2 µL DNA sample to a final volume of 22 µL. Approximately 20,000 nanoliter-sized droplets were generated from the mixture using an automated droplet generator (QX200
TM AutoDG ddPCR system (Bio-Rad, Hercules, CA, USA)), as shown in
Figure 1. The plate containing the resultant droplets was then heat-sealed with a pierceable aluminum foil using a PX1 PCR plate sealer (Bio-Rad, Hercules, CA, USA) set to run at 180 °C for 5 sec before being loaded into a C100 Touch
TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) for amplification. The thermal cycling conditions with a ramp rate of 2 °C/s at every step were set to run for 10 min enzyme activation at 95 °C, followed by 40 cycles of denaturation at 94 °C for 30 s and 1 min annealing/extension at 60 °C, enzyme deactivation step set at 98 °C for 10 min, and a final hold step at 4 °C for an infinite time period. After thermal cycling, the plate containing amplified droplets was placed in a QX200
TM Droplet Reader (Bio-Rad, Hercules, CA, USA) for reading. Prior to reading, the QuantaSoft™ software was opened on a personal computer and wells containing the samples were labeled with the appropriate reading conditions before being run.
2.8. Intra- and Inter-Assay Variability and Reproducibility of Simplex and Duplex Assays
To test the intra-assay variability, six replicates of M. bovis BCG and Mtb. H37Ra were compared using simplex and duplex assays. After testing, the resultant mean and SD of the replicates data were compared to get the percentage coefficient of variability (%CV). The copies of the two genes were directly recorded for simplex assays. But for the duplex assay, further analysis using the threshold tool was conducted to estimate the exact concentration of the two target genes within the duplex assay. The threshold tool was placed slightly above the IS1081 droplets to get the concentration of the IS6110 gene. This was subtracted from the total concentration of the two targets after placing the threshold tool slightly above the negative droplets to get the copies of the IS1081 gene, i.e., total copies of two targets (threshold line slightly above negative droplets) − IS6110 copies (threshold line slightly above IS1081 gene) = IS1081 copies. To further test if there was a difference in copies between the simplex and duplex assays (inter-assay variability) after using the threshold tool, the mean of means of each target was compared and SD of each result obtained. This was further used to calculate the %CV between the simplex and duplex assay.
2.9. Data Analysis
Real-time PCR results Cq values were generated and analyzed using the Bio-Rad CFX Manager software version 2.1. For ddPCR data, QuantaSoft™ analysis software version 1.7.4 (Bio-Rad) was used to analyze the resultant droplets. Based on thresholding tools provided by the software, proportions of positive and negative droplets were discriminated by applying the threshold line above the negative droplets. For each reaction on ddPCR, the number of accepted droplets was checked, if the droplets were ≥10,000, the results were used for analysis or else discarded. Further statistical analysis was done using the GraphPad Prism Version 6.01 software.
4. Discussion
Recent reviews by Nyaruaba et al. [
2] and Li et al. [
4] highlight a paucity in publications on the applications of ddPCR in detecting
Mtb. Less than 10 publications exist on the application of ddPCR towards TB detection and diagnosis [
2], most of which are aimed at absolute quantification of TB using simplex and duplex double dye assays. From all these studies, however, none have highlighted the use of a single dye duplex ddPCR assay combining the two genes
IS6100 and
IS1081 in TB quantification. Furthermore, none have targeted the
IS1081 gene for the detection or quantification of tuberculosis using ddPCR. Inclusion of the
IS1081 target gene in our study was of key novel importance as the gene was used before to detect rare cases of
M. tuberculosis strains where the
IS6110 target was found missing in clinical isolates [
9,
13]. This study not only aims at the inclusion of the two target genes for detecting
Mtb but also highlights the steps toward the development and evaluation of a single dye duplex ddPCR assay that can reliably distinguish the two genes within a single sample.
The Bio-Rad QX100/QX200™ Droplet Digital™ PCR system reader was designed to detect duplex targets in two separate channels (FAM and VIC/HEX) when TaqMan hydrolysis probes are used for duplex/multiplex experiments. This means that targets for duplex/multiplex reactions were to be labeled with separate dyes and then detected in separate channels during analysis. For single channel duplexing, DNA-binding dye chemistry was used and steps toward optimization and analysis described [
14]. Due to the lack of existing standards to perform, design, and analyze single dye, TaqMan-based duplex ddPCR assays, we sought to test various parameters and show the steps toward the development of such an assay suitable for detecting
Mtb samples.
Why
Mtb. H37Ra and
M. bovis BCG?
Mtb. H37Ra and
M. bovis BCG were chosen for optimization and evaluation of the assay since these two bacteria have known copies of the target genes to be quantified. From existing literature,
Mtb. H37Ra are shown to have 17 complete copies of the
IS6110 gene, a slight difference from the 16 copies observed in the
Mtb. H37Rv strain [
15] and 5 copies of the
IS1081 gene.
M. bovis BCG, on the other hand, was documented to have 1 copy of
IS6110 and 5 copies of the
IS1081 gene [
10]. Due to these facts, one can visually approximate the location of target droplet genes within the duplex assay, as shown in
Figure 6. Having samples with known copies of the target genes to be tested in a duplex assay can help locate the genes within the duplex assay with ease. The higher the copy numbers of a particular gene, the higher the concentrations of droplets of that gene within a particular duplex assay.
Sample concentration plays an important role in the determination of positive, negative, double positive, and double negative droplet partitions. From our results in
Figure 5, we have clearly shown that the concentration of droplets in a highly concentrated duplex single dye assay will be located in the double positive droplets and not within the individual targets as shown in simplex assays. Highly concentrated samples that were not digested/diluted will produce only single amplitudes that cannot be quantified as either positive or negative automatically by the QuantaSoft™ software. For accurate determination and quantification of droplets, dilutions of the sample to lesser concentrations will help to determine the correct position of positive droplets. Additionally, sample volume can also be adjusted after establishing the concentration of the sample. Most commercial detection assays use higher sample volumes; we believe that after the sample concentration is determined, the volume can be adjusted (e.g., 5 µL if the concentration is low or 2 µL if the concentration is high) to achieve optimum detection, quantification, and/or droplet separation results.
Figure 8 is a schematic representation of how droplets separate in a duplex assay containing highly and lowly concentrated samples.
From pre-existing literature, varying primer and probe concentrations were used to distinguish target genes within a single channel using the same dye [
16,
17]. However, from our study, varying this concentration was shown to have very little effect on the separation of droplets within our duplex assay. This variation of results was thought to be due to differences in the efficiency of amplification for each of the targets—the lower target (
IS1081) might have not been amplified so efficiently as the upper target (
IS6110). Although it is a probe-based assay, the length of amplicons could also have some effect on amplitude (due to the longer time needed to amplify longer amplicons, making it less efficient in comparison to others).
The sensitivity and specificity of both the ddPCR and qPCR assays in this experiment were equal (100%). This is mostly due to the fact that the samples determined were cultured samples and not clinical isolates. We believe that this sensitivity and specificity may vary if clinical isolates are used. Especially in cases where the concentration of targets is low, more studies need to be conducted on clinical samples comparing the sensitivity and specificity of the assay to commonly used methods like sputum smear microscopy and the Xpert
® MTB/RIF ULTRA assay. Several studies [
2], however, established the sensitivity and specificity of ddPCR in detection of
M. tuberculosis from various clinical isolates and compared this sensitivity and specificity to other techniques like qPCR. From their findings, ddPCR was superior to qPCR in quantifying low targets from plasma [
18,
19,
20] and sputum [
3].
Prospectively, we think that duplex target detection and quantification using the same dye gives room for higher multiplexing and saves on the high costs incurred in simplex experiments—a technique not possible by qPCR. Using the HEX channel, one may include one or two extra targets (triplex or fourplex assay). This assay may also be used in future studies to establish if the two genes have a direct role in the pathogenesis or severity of TB infections. The assay may also be explored further in the future to test the effect of drugs on the two targets. Mass screening of a large number of patient samples may be done using this assay to also establish the epidemiology and distribution of the two targets within a population. Lastly, since our tests were mostly conducted on cultured samples; it would be of great benefit for future experiments to compare the assay’s superiority against other molecular methods like MALDI–TOF mass spectrometry.