Bioremediation of Historically Chlorimuron-Ethyl-Contaminated Soil by Co-Culture Chlorimuron-Ethyl-Degrading Bacteria Combined with the Spent Mushroom Substrate
Abstract
1. Introduction
2. Materials and methods
2.1. Bacterial Strains
2.2. Chemicals and Media
2.3. Soil Samples and SMS
2.4. The Chlorimuron-Ethyl Extraction Assay
2.5. HPLC Instrumentation and Conditions
2.6. Effects of Laccase Crude Extract from SMS on the Degradation of Chlorimuron-Ethyl and Carrier Selection
2.7. Degradation of Chlorimuron-Ethyl by Sterilized P. eryngiu-SMS and Unsterilized P. eryngiu-SMS
2.8. Preparation of the P. eryngiu-SMS-CB
2.9. Degradation of Chlorimuron-Ethyl by P. eryngiu-SMS, Co-Culture Bacteria and P. eryngiu-SMS-CB
2.10. Remediation of Chlorimuron-Ethyl-Contaminated Soil
2.11. Plant Culture
2.12. Determination of SOD Activity and SP
2.13. Data Analysis and Statistics
3. Results and Discussion
3.1. Degradation of Chlorimuron-Ethyl by Crude Extracts Containing Laccase from SMS and Carrier Selection
3.2. Degradation of Chlorimuron-Ethyl by Sterilized P. eryngiu-SMS and Unsterilized P. eryngiu-SMS
3.3. Preparation of the P. eryngiu-SMS-CB
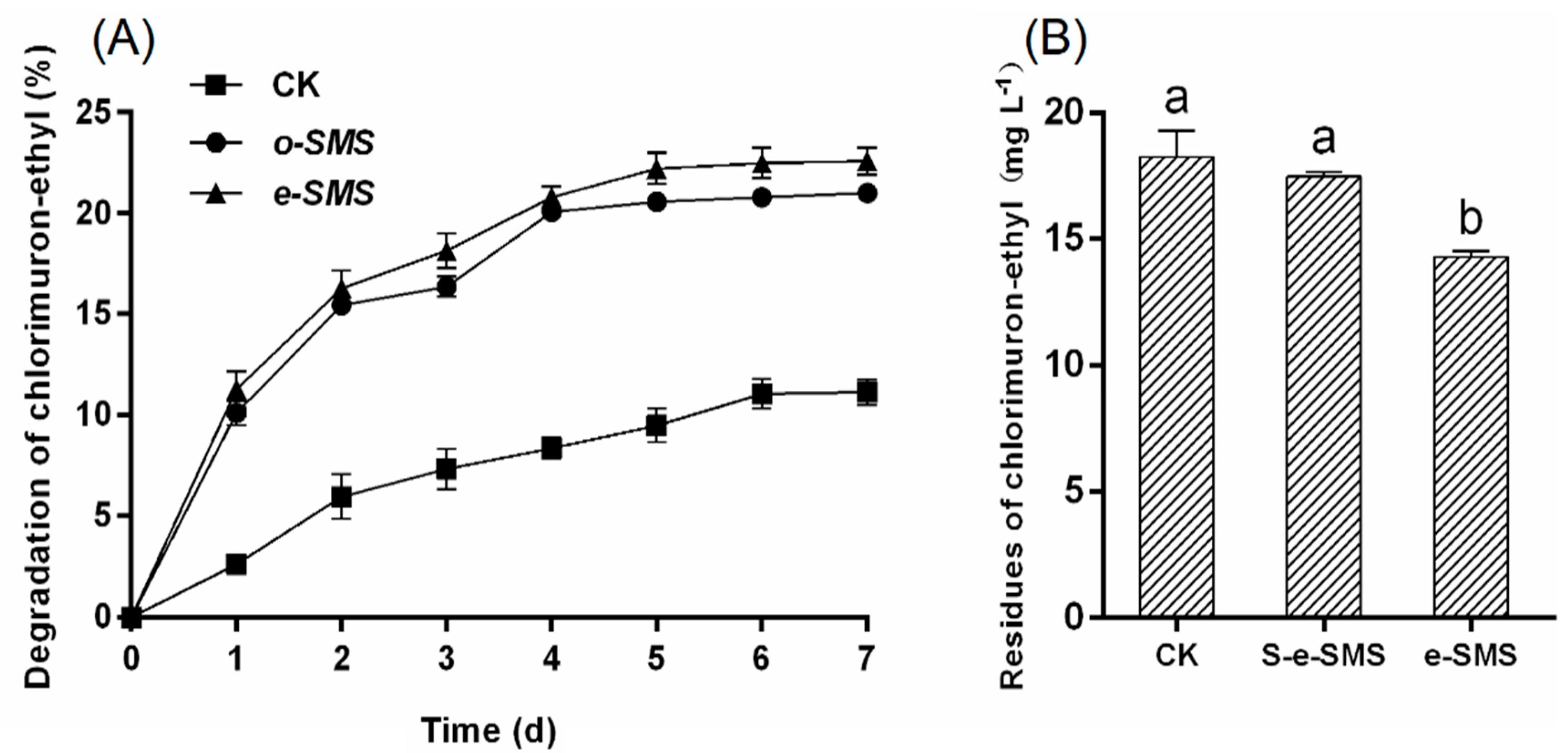
3.4. Degradation of Chlorimuron-Ethyl by SMS, Co-Culture Bacteria and P. eryngiu-SMS-CB
3.5. Remediation of Chlorimuron-Ethyl-Contaminated Soil by the P. eryngiu-SMS-CB
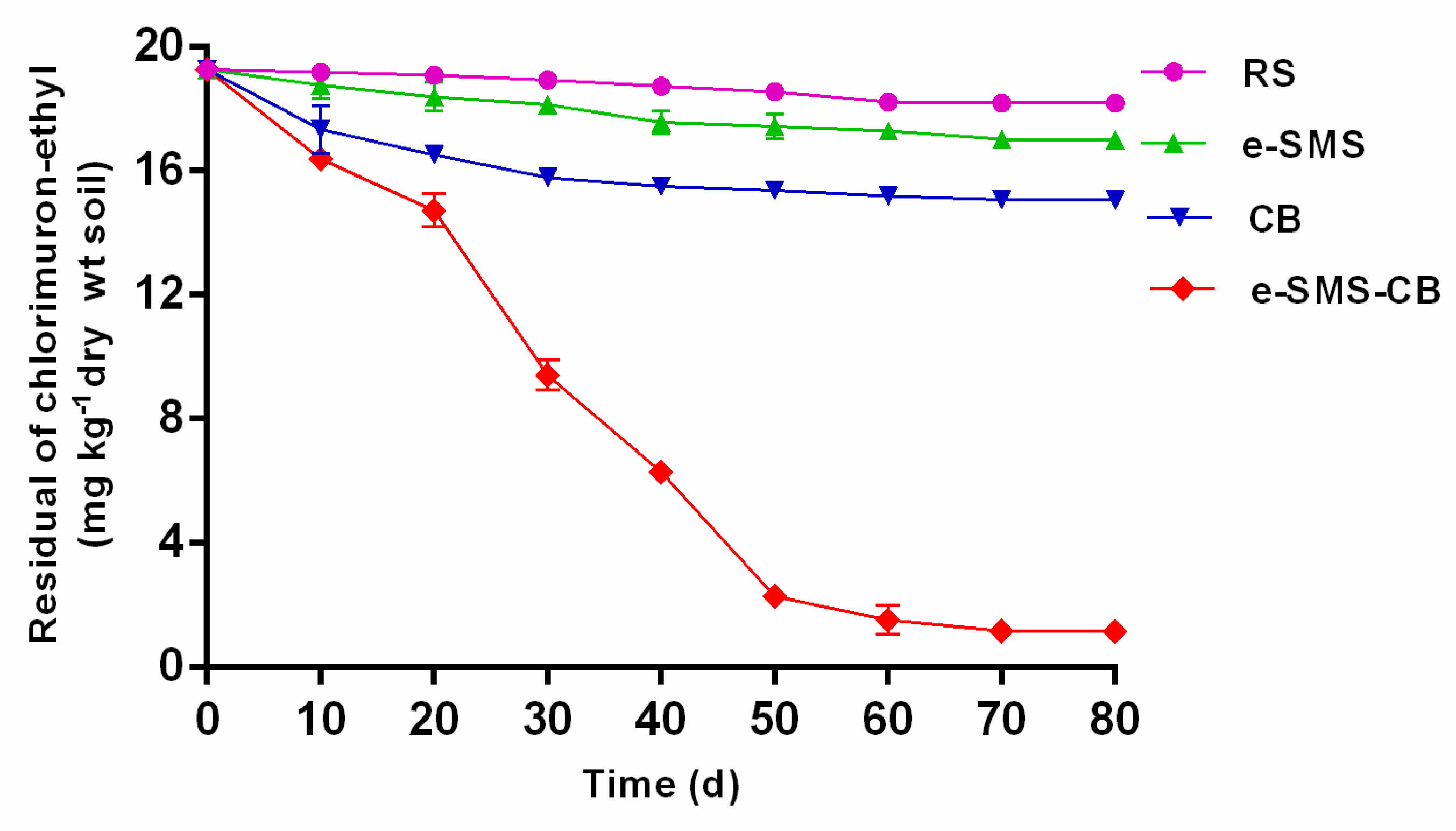
3.5.1. Effects of the P. eryngiu-SMS-CB dosage on Chlorimuron-Ethyl Degradation in Soil
3.5.2. Remediation of Chlorimuron-Ethyl-Contaminated Soil
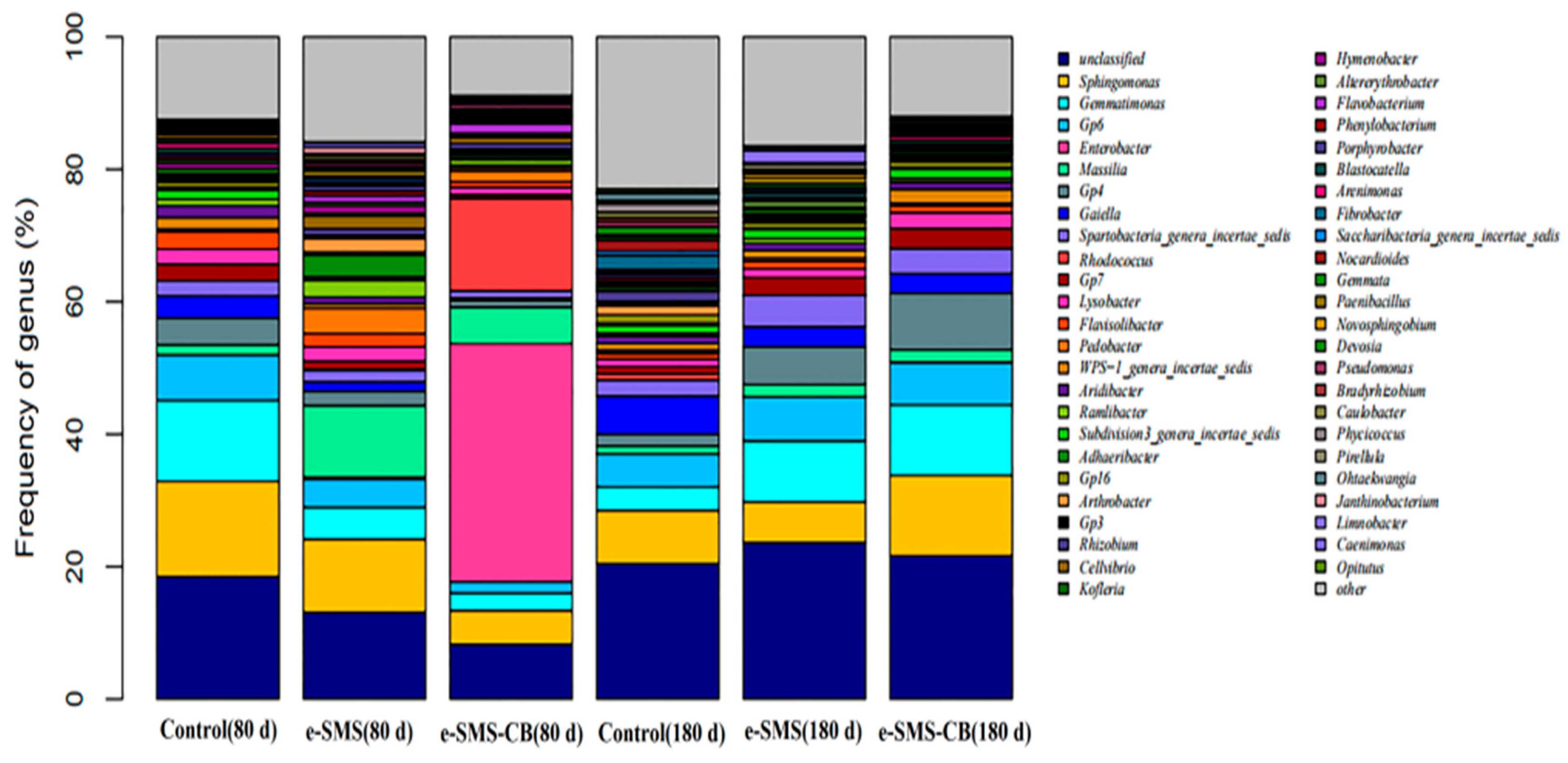
3.5.3. Bacterial Diversity and Community Structure in Soil
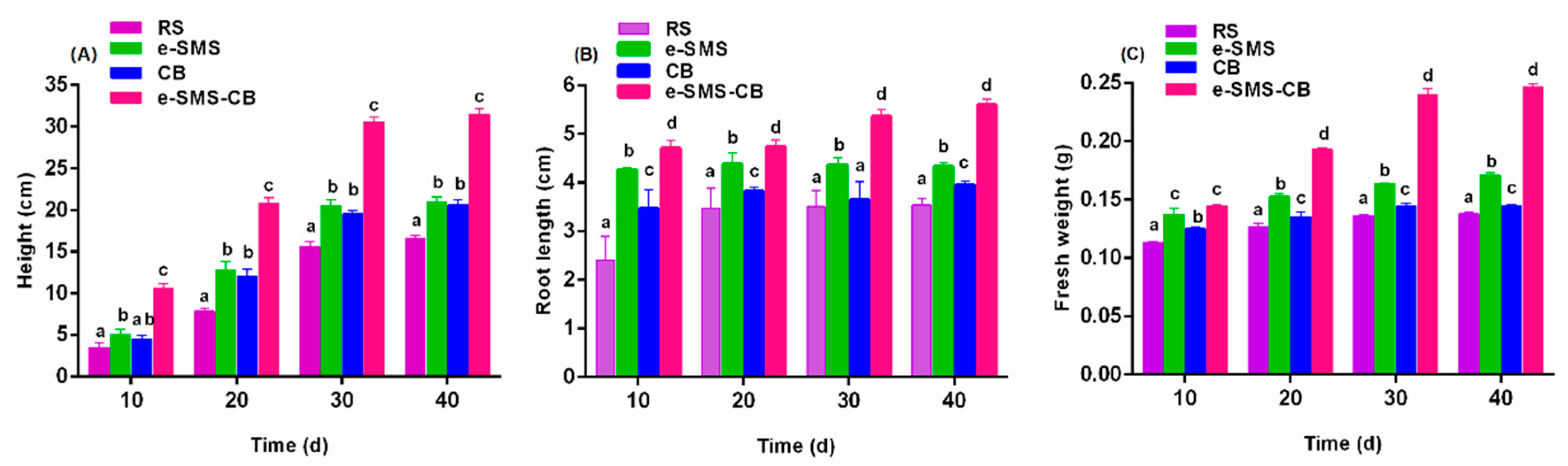
3.5.4. Plant Culture
3.5.5. SOD and SP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xiong, M.H.; Hu, Z.G.; Zhang, Y.; Cheng, X.S.; Li, C.Y. Survival of GFP-tagged Rhodococcus sp. D310-1 in chlorimuron-ethyl-contaminated soil and its effects on the indigenous microbial community. J. Hazard. Mater. 2013, 252, 347–354. [Google Scholar] [CrossRef]
- Zhang, X.L.; Zhang, H.W.; Li, X.; Su, Z.C.; Wang, J.J.; Zhang, C.G. Isolation and characterization of Sporobolomyces sp. LF1 capable of degrading chlorimuron-ethyl. J. Environ. Biol. 2009, 21, 1253–1260. [Google Scholar] [CrossRef]
- Ren, W.J.; Zhou, Q.X.; Wang, M.; Jadeja, R.N. Interactive effects of chlorimuron-ethyl and copper(II) on their sorption and desorption on two typical Chinese soils. Eur. J. Soil Sci. 2011, 62, 882–890. [Google Scholar] [CrossRef]
- Al-Kharusi, S.; Abed, R.M.M.; Dobretsov, S. Changes in respiration activities and bacterial communities in a bioaugmented oil-polluted soil in response to the addition of acylhomoserine lactones. Int. Biodeterior. Biodegrad. 2016, 107, 165–173. [Google Scholar] [CrossRef]
- Xie, X.H.; Liu, N.; Ping, J.; Zhang, Q.Y.; Zheng, X.L.; Liu, J.S. Illumina MiSeq sequencing reveals microbial community in HA process for dyeing wastewater treatment fed with different co-substrates. Chemosphere 2018, 201, 578–585. [Google Scholar] [CrossRef]
- Yang, L.Q.; Li, X.Y.; Li, X.; Su, Z.C.; Zhang, C.G.; Zhang, H.W. Bioremediation of chlorimuron-ethyl-contaminated soil by Hansschlegelia sp. strain CHL1 and the changes of indigenous microbial population and N-cycling function genes during the bioremediation process. J. Hazard. Mater. 2014, 274, 314–321. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.H.; Mu, W.H.; Wang, J.X.; Pan, H.Y.; Li, Y. Biodegradation of chlorimuron-ethyl by the bacterium Klebsiella jilinsis 2N3. Environ. Sci. Health Part B 2010, 45, 501–507. [Google Scholar] [CrossRef]
- Zang, H.L.; Yu, Q.; Lv, T.Y.; Cheng, Y.; Feng, L.; Cheng, X.S.; Li, C.Y. Insights into the degradation of chlorimuron-ethyl by Stenotrophomonas maltophilia D310-3. Chemosphere 2016, 144, 176–184. [Google Scholar] [CrossRef]
- Li, C.Y.; Lv, T.Y.; Liu, W.J.; Zang, H.L.; Cheng, Y.; Li, D.P. Efficient degradation of chlorimuron-ethyl by a bacterial consortium and shifts in the aboriginal microorganism community during the bioremediation of contaminated-soil. Ecotoxicol. Environ. Saf. 2017, 139, 423–430. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Parra-Saldivar, R.; Hu, H.B.; Wang, W.; Zhang, X.H.; Iqbal, H.M.N. Immobilized ligninolytic enzymes: An innovative and environmental responsive technology to tackle dye-based industrial pollutants—A review. Sci. Total Environ. 2017, 576, 646–659. [Google Scholar] [CrossRef]
- Medina, E.; Paredes, C.; Bustamante, M.A.; Moral, R.; Moreno-Caselles, J. Relationships between soil physico-chemical, chemical and biological properties in a soil amended with spent mushroom substrate. Geoderma 2012, 173, 152–161. [Google Scholar] [CrossRef]
- Peregrina, F.; Larrieta, C.; Colina, M.; Mariscal-Sancho, I.; Martin, I.; Martinez-Vidaurre, J.M.; Garcia-Escudero, E. Spent mushroom substrates influence soil quality and nitrogen availability in a semiarid vineyard soil. Soil Sci. Soc. Am. J. 2012, 76, 1655–1666. [Google Scholar] [CrossRef]
- Meng, L.Q.; Zhang, S.M.; Gong, H.N.; Zhang, X.C.; Wu, C.D.; Li, W.G. Improving sewage sludge composting by addition of spent mushroom substrate and sucrose. Bioresour. Technol. 2018, 253, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Marínbenito, J.M.; Herrerohernández, E.; Andrades, M.S. Effect of different organic amendments on the dissipation of linuron, diazinon and myclobutanil in an agricultural soil incubated for different time periods. Sci. Total Environ. 2014, 476–477, 611–621. [Google Scholar] [CrossRef]
- Singh, A.; Abdullah, N.; Vikineswary, S. Optimization of extraction of bulk enzymes from spent mushroom compost. J. Chem. Technol. Biotechnol. 2003, 78, 743–752. [Google Scholar] [CrossRef]
- Zeng, S.Q.; Qin, X.L.; Xia, L.M. Degradation of the herbicide isoproturon by laccase-mediator systems. Biochem. Eng. J. 2017, 119, 92–100. [Google Scholar] [CrossRef]
- Li, C.Y.; Zang, H.L.; Yu, Q.; Lv, T.Y.; Cheng, Y.; Cheng, X.S.; Liu, K.R.; Liu, W.J.; Xu, P.P.; Lan, C.Z. Biodegradation of chlorimuron-ethyl and the associated degradation pathway by Rhodococcus sp. D310-1. Environ. Sci. Pollut. Res. 2016, 23, 8794–8805. [Google Scholar] [CrossRef]
- Li, C.Y.; Cao, H.M.; An, X.J. Isolation and identifification of a chlorimuron-ethyl degrading bacterium and optimization of its degradation conditions. J. Northeast Agric. Univ. 2016, 47, 65–72. [Google Scholar]
- Zang, H.L.; Wang, H.L.; Miao, L.; Cheng, Y.; Zhang, Y.T.; Liu, Y.; Sun, S.S.; Wang, Y.; Li, C.Y. Carboxylesterase, a de-esterification enzyme, catalyzes the degradation of chlorimuron-ethyl in Rhodococcus erythropolis D310-1. J. Hazard. Mater. 2019, 387, 121684. [Google Scholar] [CrossRef]
- Wei, Y.Q.; Zhao, Y.; Zhao, X.Y.; Gao, X.T.; Zheng, Y.S.; Zuo, H.D.; Wei, Z.M. Roles of different humin and heavy-metal resistant bacteria from composting on heavy metal removal. Bioresour. Technol. 2020, 296, 122375. [Google Scholar] [CrossRef]
- Ben Mefteh, F.; Frikha, F.; Daoud, A.; Bouket, A.C.; Luptakova, L.; Alenezi, F.N.; Al-Anzi, B.S.; Oszako, T.; Gharsallah, N.; Belbahri, L. Response surface methodology optimization of an acidic protease produced by Penicillium bilaiae Isolate TDPEF30, a newly recovered endophytic fungus from healthy roots of date palm trees (Phoenix dactylifera L.). Microorganisms 2019, 7, 74. [Google Scholar] [CrossRef]
- Ahmad, M.; Yang, Q.S.; Zhang, Y.Y.; Ling, J.; Sajjad, W.; Qi, S.H.; Zhou, W.G.; Zhang, Y.; Lin, X.C.; Zhang, Y.H.; et al. The distinct response of phenanthrene enriched bacterial consortia to different PAHs and their degradation potential: A mangrove sediment microcosm study. J. Hazard. Mater. 2019, 380, 120863. [Google Scholar] [CrossRef] [PubMed]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.L.; Rizwan, M.; Li, M.; Song, F.R.; Zhou, S.J.; He, X.; Ding, R.; Dai, Z.H.; Yuan, Y.; Cao, M.H.; et al. Comparative efficacy of organic and inorganic silicon fertilizers on antioxidant response, Cd/Pb accumulation and health risk assessment in wheat (Triticum aestivum L.). Environ. Pollut. 2019, 255, 113146. [Google Scholar] [CrossRef] [PubMed]
- Seth, K.; Aery, N.C. Boron induced changes in biochemical constituents, enzymatic activities, and growth performance of wheat. Acta Physiol. Plant. 2015, 39, 244. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.J.; Ng, T.B.; Deng, X.Z.; Lin, J.; Ye, X.Y. Laccases: Production, expression regulation, and applications in pharmaceutical biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef]
- Fernández-Fernández, M.; Sanromán, M.A.; Moldes, D. Recent developments and applications of immobilized laccase. Biotechnol. Adv. 2013, 31, 1808–1825. [Google Scholar]
- Ba, S.; Arsenault, A.; Hassani, T.; Jones, J.P.; Cabana, H. Laccase immobilization and insolubilization: From fundamentals to applications for the elimination of emerging contaminants in wastewater treatment. Crit. Rev. Biotechnol. 2013, 33, 404–418. [Google Scholar] [CrossRef]
- Cheng, Z.W.; Lu, L.C.; Kennes, C.; Ye, J.X.; Yu, J.M.; Chen, D.Z.; Chen, J.M. A composite microbial agent containing bacterial and fungal species: Optimization of the preparation process, analysis of characteristics, and use in the purification for volatile organic compounds. Bioresour. Technol. 2016, 218, 751–760. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, C.; Wen, Y.Z.; Liu, W.P. Enantioselective separation and degradation of the herbicide dichlorprop methyl in sediment. Chirality 2009, 21, 480–483. [Google Scholar] [CrossRef]
- Jaswal, R.; Pathak, A.; Chauhan, A. Metagenomic evaluation of bacterial and fungal assemblages enriched within diffusion chambers and microbial traps containing uraniferous soils. Microorganisms 2019, 7, 324. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Si, Y.X.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2019, 22, 10788–10799. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Li, Y.; Sun, R.X.; Lin, Y.L.; Yu, H.; Xue, Y.W.; Zhou, X.Z.; Liu, W.X.; Yan, L.L.; et al. The drivers of bacterial community underlying biogeographical pattern in Mollisol area of China. Ecotoxicol. Environ. Saf. 2019, 177, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Sellier, Y.; Marliot, F.; Bessieres, B.; Stirnemann, J.; Encha-Razavi, F.; Guilleminot, T.; Haicheur, N.; Pages, F.; Ville, Y.; Leruez-Ville, M. Adaptive and innate immune cells in fetal human cytomegalovirus-infected brains. Microorganisms 2020, 8, 176. [Google Scholar] [CrossRef]
- López-Gonzalez, J.A.; Suarez-Estrella, F.; Vargas-Garcia, M.C.; Lopez, M.J.; Jurado, M.M.; Moreno, J. Dynamics of bacterial microbiota during lignocellulosic waste composting: Studies upon its structure, functionality and biodiversity. Bioresour. Technol. 2015, 175, 406–416. [Google Scholar] [CrossRef]
- Kawanobe, M.; Toyota, K.; Fujita, T.; Hatta, D. Evaluation of nematicidal activity of fluensulfone against non-target free-living nematodes under field conditions. Agronomy 2019, 9, 853. [Google Scholar] [CrossRef]
- Garcia-Delgado, C.; D’Annibale, A.; Pesciaroli, L.; Yunta, F.; Crognale, S.; Petruccioli, M.; Eymar, E. Implications of polluted soil biostimulation and bioaugmentation with spent mushroom substrate (Agaricus bisporus) on the microbial community and polycyclic aromatic hydrocarbons biodegradation. Sci. Total Environ. 2015, 508, 20–28. [Google Scholar] [CrossRef]
- Min, J.; Wang, B.; Hu, X.K. Effect of inoculation of Burkholderia sp. strain SJ98 on bacterial community dynamics and para-nitrophenol, 3-methyl-4-nitrophenol, and 2-chloro-4-nitrophenol degradation in soil. Sci. Rep. 2015, 7, 5983. [Google Scholar] [CrossRef]
- Courtney, R.G.; Mullen, G.J. Soil quality and barley growth as influenced by the land application of two compost types. Bioresour. Technol. 2008, 99, 2913–2918. [Google Scholar] [CrossRef]
- Hackett, R. Spent mushroom compost as a nitrogen source for spring barley. Nutr. Cycl. Agroecosyst. 2015, 102, 253–263. [Google Scholar] [CrossRef]
- Jia, H.S.; Han, Y.Q.; Li, D.Q. Photoinhibition and active oxygen species production in detached apple leaves during dehydration. Photosynthetica 2003, 41, 151–156. [Google Scholar] [CrossRef]
- Wu, F.H.; An, Y.Q.; An, Y.R.; Wang, X.J.; Cheng, Z.Y.; Zhang, Y.; Hou, X.W.; Chen, C.X.; Wang, L.; Bai, J.G. Acinetobacter calcoaceticus CSY-P13 mitigates stress of ferulic and p-hydroxybenzoic acids in cucumber by affecting antioxidant enzyme activity and soil bacterial community. Front. Microbiol. 2018, 9, 1262. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zang, H.; Liu, W.; Cheng, Y.; Wang, H.; An, X.; Sun, S.; Wang, Y.; Hou, N.; Cui, C.; Li, C. Bioremediation of Historically Chlorimuron-Ethyl-Contaminated Soil by Co-Culture Chlorimuron-Ethyl-Degrading Bacteria Combined with the Spent Mushroom Substrate. Microorganisms 2020, 8, 369. https://doi.org/10.3390/microorganisms8030369
Zang H, Liu W, Cheng Y, Wang H, An X, Sun S, Wang Y, Hou N, Cui C, Li C. Bioremediation of Historically Chlorimuron-Ethyl-Contaminated Soil by Co-Culture Chlorimuron-Ethyl-Degrading Bacteria Combined with the Spent Mushroom Substrate. Microorganisms. 2020; 8(3):369. https://doi.org/10.3390/microorganisms8030369
Chicago/Turabian StyleZang, Hailian, Wanjun Liu, Yi Cheng, Hailan Wang, Xuejiao An, Shanshan Sun, Yue Wang, Ning Hou, Chunyu Cui, and Chunyan Li. 2020. "Bioremediation of Historically Chlorimuron-Ethyl-Contaminated Soil by Co-Culture Chlorimuron-Ethyl-Degrading Bacteria Combined with the Spent Mushroom Substrate" Microorganisms 8, no. 3: 369. https://doi.org/10.3390/microorganisms8030369
APA StyleZang, H., Liu, W., Cheng, Y., Wang, H., An, X., Sun, S., Wang, Y., Hou, N., Cui, C., & Li, C. (2020). Bioremediation of Historically Chlorimuron-Ethyl-Contaminated Soil by Co-Culture Chlorimuron-Ethyl-Degrading Bacteria Combined with the Spent Mushroom Substrate. Microorganisms, 8(3), 369. https://doi.org/10.3390/microorganisms8030369



