Inactivation of the pgmA Gene in Streptococcus mutans Significantly Decreases Biofilm-Associated Antimicrobial Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Transformation of S. mutans
2.3. Screening of a S. mutans Transposon Library for Mutants Impaired in Biofilm-Associated Gentamicin Tolerance
2.4. Determination of Minimum Bactericidal Concentration for Biofilm Cells (MBC-B) and Minimum Bactericidal Concentration for Planktonic Cells (MBC-P)
2.5. Time-Kill Assay
2.6. Identification of Interrupted Genes by Arbitrary Primed Polymerase Chain Reaction (PCR)
2.7. Construction of a pgmA Deletion Mutant
2.8. Construction of pgmA Complemented Strain
2.9. Crystal Violet Biofilm Assay
2.10. CLSM and Airyscan
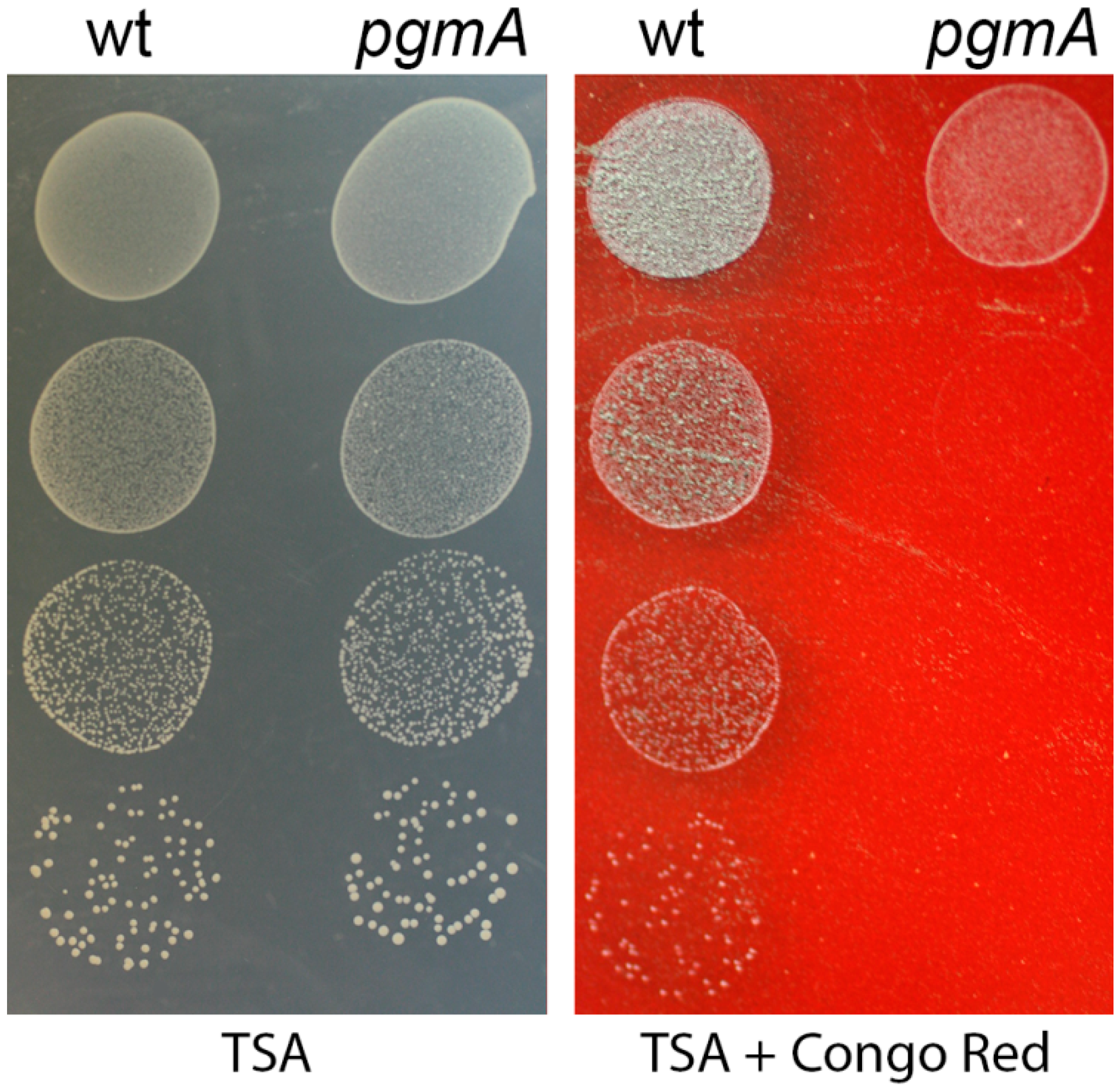
2.11. Congo Red Susceptibility Assay
2.12. Statistics
3. Results
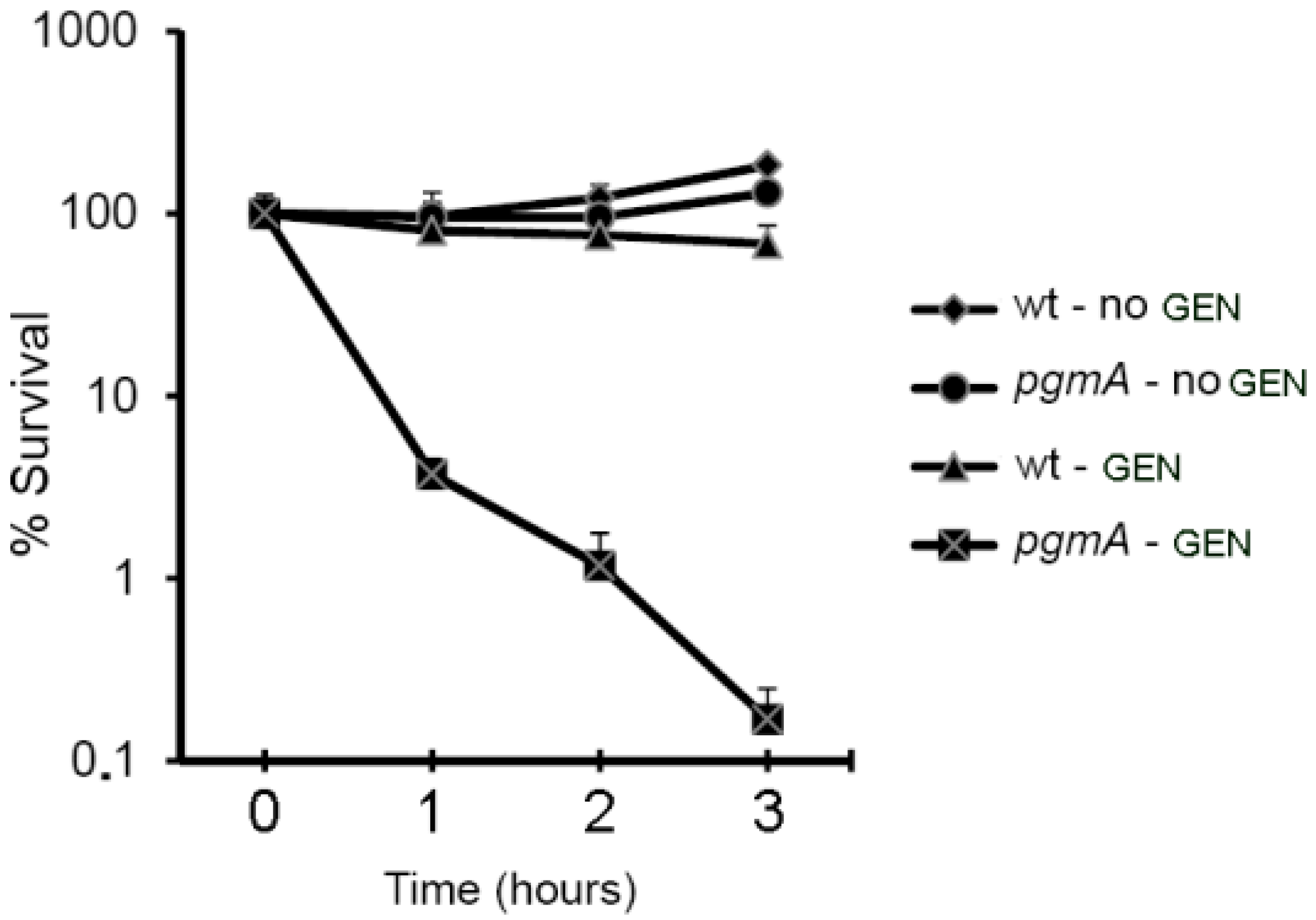
3.1. S. mutans pgmA Mutants Display Reduced Biofilm-Associated Antimicrobial Tolerance
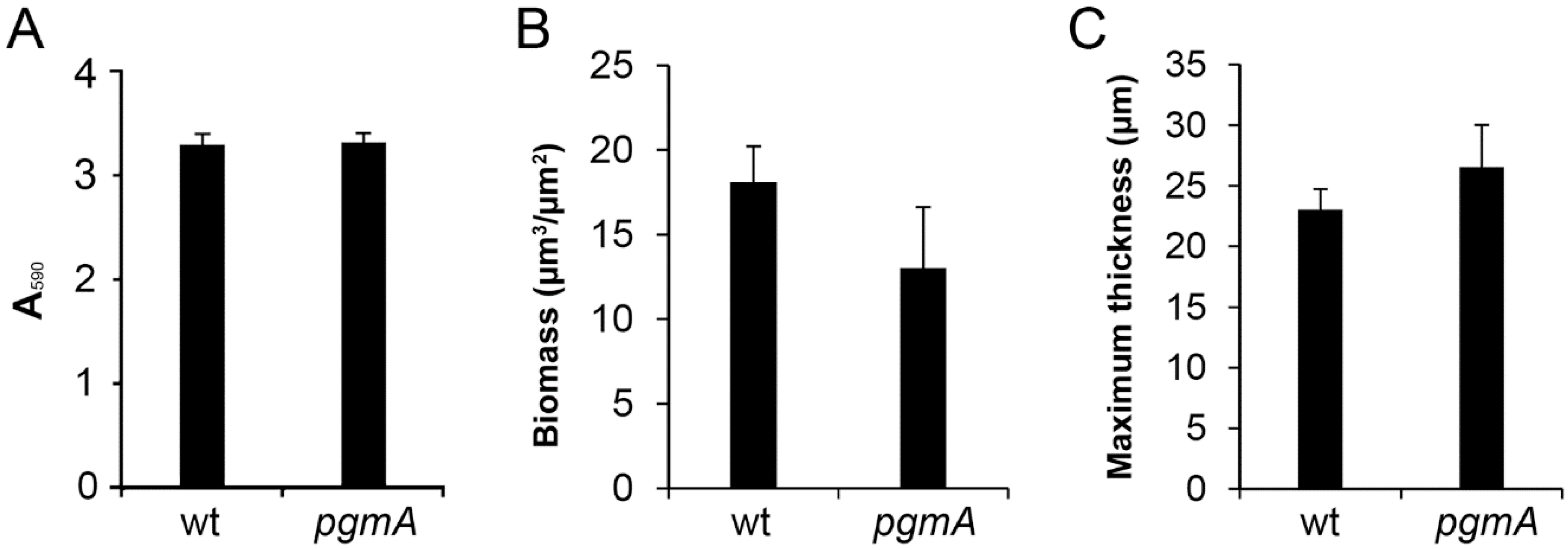
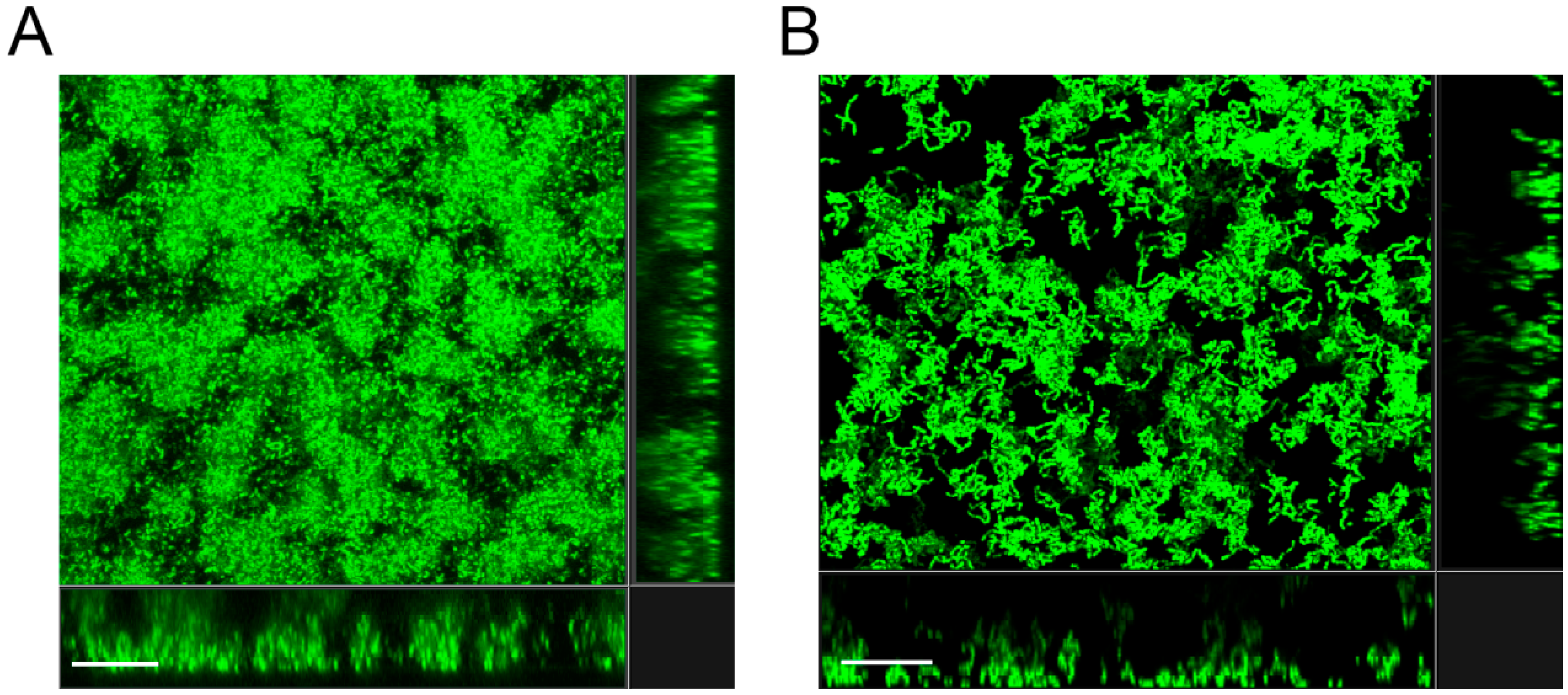
3.2. The Amount of Biofilm Formed by the S. mutant pgmA Mutant is Similar to that of the Wild Type
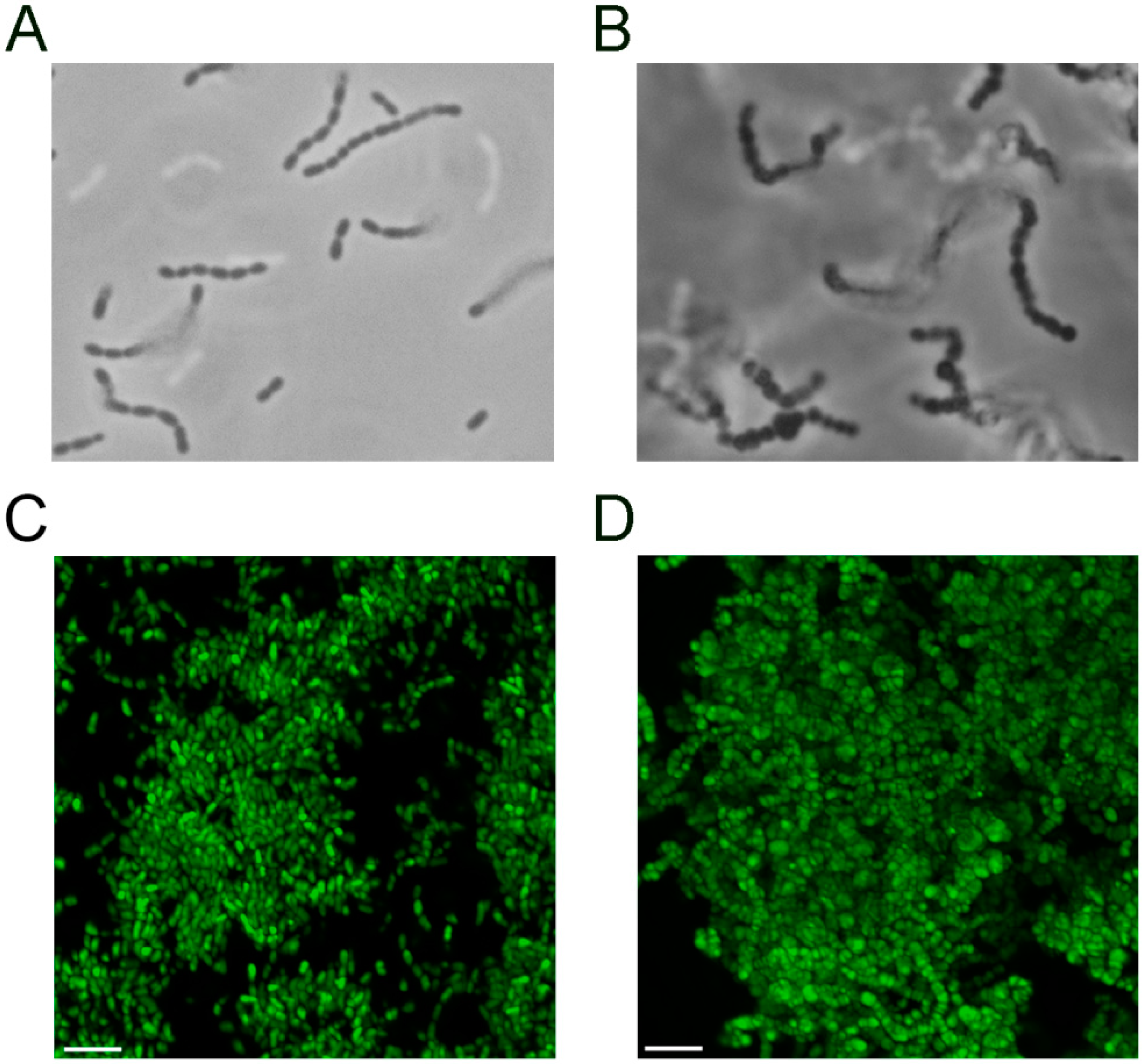
3.3. The S. mutans pgmA Mutant has Altered Cell Wall Properties
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takahashi, N.; Nyvad, B. Caries ecology revisited: Microbial dynamics and the caries process. Caries Res. 2008, 42, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Krzysciak, W.; Jurczak, A.; Koscielniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef] [PubMed]
- Schilling, K.M.; Bowen, W.H. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 1992, 60, 284–295. [Google Scholar] [PubMed]
- Colby, S.M.; Russell, R.R.B. Sugar metabolism by mutans streptococci. J. Appl. Microbiol. 1997, 83, 80S–88S. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Szilagyi, G.; Tanowitz, H.B.; Luftschein, S.; Baum, S.G. Infective endocarditis caused by Streptococcus mutans. A complication of idiopathic hypertrophic subaortic stenosis. Arch. Intern. Med. 1977, 137, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Parahitiyawa, N.B.; Jin, L.J.; Leung, W.K.; Yam, W.C.; Samaranayake, L.P. Microbiology of odontogenic bacteremia: Beyond endocarditis. Clin. Microbiol. Rev. 2009, 22, 46–64. [Google Scholar] [CrossRef]
- Bedran, T.B.L.; Azelmat, J.; Spolidorio, D.P.; Grenier, D. Fibrinogen-induced streptococcus mutans biofilm formation and adherence to endothelial cells. Biomed. Res. Int. 2013, 2013, 431465. [Google Scholar]
- Nomura, R.; Naka, S.; Nemoto, H.; Inagaki, S.; Taniguchi, K.; Ooshima, T.; Nakano, K. Potential involvement of collagen-binding proteins of Streptococcus mutans in infective endocarditis. Oral Dis. 2013, 19, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Beg, A.M.; Jones, M.N.; Miller-Torbert, T.; Holt, R.G. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem. Biophys. Res. Commun. 2002, 298, 75–79. [Google Scholar] [CrossRef]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response. J. Mol. Biol. 2015, 427, 3628–3645. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: From molecular biofilm biology to new treatment possibilities. APMIS Suppl. 2014, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T.; Jensen, P.O.; Wang, H.; Hoiby, N. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv. Drug. Deliv. Rev. 2015, 85. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Christiansen, N.; Hoiby, N.; Twetman, S.; Givskov, M.; Tolker-Nielsen, T. A mariner transposon vector adapted for mutagenesis in oral streptococci. Microbiologyopen 2014, 3, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Rybtke, M.; Givskov, M.; Hoiby, N.; Twetman, S.; Tolker-Nielsen, T. The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int. J. Antimicrob. Agents 2016, 48, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Jakobsen, T.H.; Givskov, M.; Twetman, S.; Tolker-Nielsen, T. Oxidative stress response plays a role in antibiotic tolerance of Streptococcus mutans biofilms. Microbiology 2019, 165, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Ajdic, D.; McShan, W.M.; McLaughlin, R.E.; Savic, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef]
- Kessler, B.; de Lorenzo, V.; Timmis, K.N. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: Regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 1992, 233, 293–301. [Google Scholar] [CrossRef]
- LeBlanc, D.J.; Lee, L.N.; Abu-Al-Jaibat, A. Molecular, genetic, and functional analysis of the basic replicon of pVA380–1, a plasmid of oral streptococcal origin. Plasmid 1992, 28, 130–145. [Google Scholar] [CrossRef]
- Mair, R.W.; Senadheera, D.B.; Cvitkovitch, D.G. CinA is regulated via ComX to modulate genetic transformation and cell viability in Streptococcus mutans. FEMS Microbiol. Lett. 2012, 331, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Biswas, I.; Drake, L.; Johnson, S.; Thielen, D. Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. Biotechniques 2007, 42, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Rigby, K.; Lai, Y.; Nair, V.; Peschel, A.; Schittek, B.; Otto, M. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob. Agents Chemother. 2009, 53, 4200–4210. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.C.; Sung, C.K.; Lee, J.H.; Morrison, D.A.; Cvitkovitch, D.G. PCR ligation mutagenesis in transformable streptococci: Application and efficiency. J. Microbiol. Methods 2002, 49, 193–205. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersboll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- DeFrancesco, A.S.; Masloboeva, N.; Syed, A.K.; DeLoughery, A.; Bradshaw, N.; Li, G.W.; Gilmore, M.S.; Walker, S.; Losick, R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, E5969–E5978. [Google Scholar] [CrossRef]
- Boels, I.C.; Ramos, A.; Kleerebezem, M.; de Vos, W.M. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 2001, 67, 3033–3040. [Google Scholar] [CrossRef]
- Lazarevic, V.; Soldo, B.; Medico, N.; Pooley, H.; Bron, S.; Karamata, D. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 2005, 71, 39–45. [Google Scholar] [CrossRef]
- Bizzini, A.; Majcherczyk, P.; Beggah-Moller, S.; Soldo, B.; Entenza, J.M.; Gaillard, M.; Moreillon, P.; Lazarevic, V. Effects of alpha-phosphoglucomutase deficiency on cell wall properties and fitness in Streptococcus gordonii. Microbiology 2007, 153, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, J.T.; Stannard, J.A.; Lauth, X.; Ostland, V.E.; Powell, H.C.; Westerman, M.E.; Nizet, V. Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development. Infec. Immun. 2005, 73, 6935–6944. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Campbell, J.; Kim, Y.; Swoboda, J.G.; Mylonakis, E.; Walker, S.; Gilmore, M.S. Wall teichoic acid protects Staphylococcus aureus from inhibition by Congo red and other dyes. J. Antimicrob. Chemother. 2012, 67, 2143–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristian, S.A.; Datta, V.; Weidenmaier, C.; Kansal, R.; Fedtke, I.; Peschel, A.; Gallo, R.L.; Nizet, V. D-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 2005, 187, 6719–6725. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.I.; Hwang, G.; Santos, P.H.; Campanella, O.H.; Koo, H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect. Microbiol. 2015, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Tam, A.; Steinberg, D. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 2007, 153, 1307–1317. [Google Scholar] [CrossRef]
| Strains, Plasmid or Oligonucleotides | Relevant Characteristics or Sequence | Source |
|---|---|---|
| Streptococcus | ||
| S. mutans UA159 | American Type Culture Collection (ATCC 700610) | [18] |
| S. mutans UA159 pgmA-Tn | This study | |
| S. mutans UA159 pgmA | This study | |
| S. mutans UA159 pgmA pDL277 | This study | |
| S. mutans UA159 pgmA pDL277-PgmA | This study | |
| E. coli | ||
| HB101 | recA thi pro leu hsdRM, Smr; strain used for maintainance and proliferation of plasmids | [19] |
| Plasmids | ||
| pDL277 | SpecR; plasmid used for complementation | [20,21] |
| Oligonucleotides | Sequence | |
| Erm-PA | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT | |
| Erm-PB | GGCCGGCCAGTCGGCAGCGACTCATAGAAT | |
| 1077-P1 | ACTAGCTTGCGGTGATGTCG | |
| 1077-P2 | GGCGCGCCCCTCCTTGGTCTTTTCATCCATTGC | |
| 1077-P3 | GGCCGGCCTACTTCAGGTACCGAGCCGAAA | |
| 1077-P4 | GTCAGTGAATCTGTTAAAGGTGCT | |
| 1077compF | TATAGGATCCTACGGCGGCAATATCCAGAC | |
| 1077compR | TATAGGATCCTGTTGAACAAGGAAATCATAAAGAC | |
| MBC-B | Fold Change | MBC-P | Fold Change | |
|---|---|---|---|---|
| S. mutans UA159 | 300 | 12 | ||
| S. mutans UA159 pgmA-Tn | 38 | 8× | 3 | 4× |
| S. mutans UA159 pgmA | 38 | 8× | 3 | 4× |
| S. mutans UA159 pgmA pDL277 | 38 | 8× | N.D. | |
| S. mutans UA159 pgmA pDL277-PgmA | 600 | +2× | N.D. |
| Linezolid MBC-B | Fold Change | Linezolid MBC-P | Fold Change | Vancomycin MBC-B | Fold Change | Vancomycin MBC-P | Fold Change | |
|---|---|---|---|---|---|---|---|---|
| S. mutans UA159 | >50 | 2.5 | 32 | 1.25 | ||||
| S. mutans UA159 pgmA | 12.5 | >4× | 1.25 | 2× | 8 | 4x | 1.25 | 1× |
| S. mutans UA159 pgmA/pDL277 | 12.5 | >4× | N.D. | 8 | 4x | N.D. | ||
| S. mutans UA159 pgmA/pDL277-PgmA | >50 | 1× | N.D. | 32 | 1x | N.D. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nilsson, M.; Givskov, M.; Twetman, S.; Tolker-Nielsen, T. Inactivation of the pgmA Gene in Streptococcus mutans Significantly Decreases Biofilm-Associated Antimicrobial Tolerance. Microorganisms 2019, 7, 310. https://doi.org/10.3390/microorganisms7090310
Nilsson M, Givskov M, Twetman S, Tolker-Nielsen T. Inactivation of the pgmA Gene in Streptococcus mutans Significantly Decreases Biofilm-Associated Antimicrobial Tolerance. Microorganisms. 2019; 7(9):310. https://doi.org/10.3390/microorganisms7090310
Chicago/Turabian StyleNilsson, Martin, Michael Givskov, Svante Twetman, and Tim Tolker-Nielsen. 2019. "Inactivation of the pgmA Gene in Streptococcus mutans Significantly Decreases Biofilm-Associated Antimicrobial Tolerance" Microorganisms 7, no. 9: 310. https://doi.org/10.3390/microorganisms7090310






