Screening of the High-Rhizosphere Competent Limoniastrum monopetalum’ Culturable Endophyte Microbiota Allows the Recovery of Multifaceted and Versatile Biocontrol Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) Analysis of Soil Samples
2.3. Isolation of Culturable Endophytic Bacterial Microbiota from L. monopetalum Collected in Tunisia
2.4. Measurement of PGP Activities of L. monopetalum Bacterial Endophytes
2.4.1. Direct Plant Growth Promoting Rhizobacteria (PGPR) Activities
Growth on Nitrogen Free Medium
Phosphate Solubilization
Siderophores Production
Indole Acetic Acid (IAA) Production
2.4.2. Indirect PGP Activities
Screening of Isolates for Extracellular Enzyme Production
Protease Production
Gelatinase Production
Chitinase Production
Cellulase Production
Amylase Production
Pectinase Production
Glucanase Production
Hydrogen Cyanide (HCN) Production
2.5. In Vitro Antibacterial and Antifungal Assays
2.6. Antibacterial Assays
2.7. Antifungal Assays
2.8. Metal Stress Resistance of L. monopetalum Bacterial Endophytes
2.9. Antibiotic Resistance of L. monopetalum Bacterial Endophytes
2.10. Bacterial DNA Extraction, 16S-rDNA Amplification, Sequencing and Phylogenetic Analysis
2.11. Isolation of the Emerging PSC1 Olive Tree Fungal Pathogen
2.12. In Planta Pathogenicity Assays
2.13. Pathogenicity Test on Potato Tubers
2.14. Pathogenicity Assays on Olive Twigs
2.15. Effects of LMRE 36 Treatment on Fusarium solani (Fso7) Disease Severity on Potato Tubers
2.16. Effects of LMRE 36 Treatments on Fusarium solani (Fso6, and Fso7) and Fusarium sp. PSC1 Disease Severity on Olive Twigs
2.17. Statistical Analysis
3. Results
3.1. ICP-OES Analysis of Rhizosphere and Soil Samples Surrounding L. monopetalum
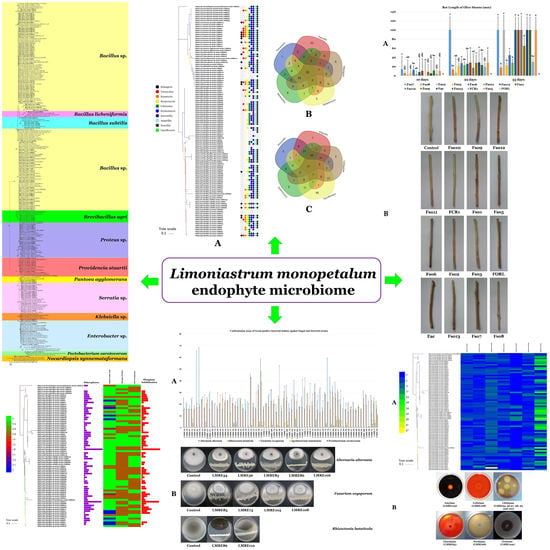
3.2. Isolation of L. monopetalum Bacterial Endophytic Microbiota
3.3. PGP Potential of L. monopetalum Bacterial Endophytes
3.3.1. Direct PGP Activities
3.3.2. Indirect PGP Activities
3.4. Biocontrol Ability of L. monopetalum Bacterial Endophytes Towards Relevant Bacterial and Fungal Plant Pathogens
3.5. Resistance of L. monopetalum Bacterial Endophytic Communities to Antibiotics and Metals
3.6. Biocontrol Ability of L. monopetalum Bacterial Endophyte LMRE 36 Towards Fusarium spp. Plant Pathogens
3.6.1. Effects of LMRE 36 Treatments on F. solani Fso7 Disease Severity on Potato Tubers
3.6.2. Effects of LMRE 36 Treatments on F. solani Fso7 Disease Severity on Olive Twigs
3.7. Isolation of the Emerging PSC1 Olive Tree Fungal Pathogen
3.8. Biocontrol Ability and Effects on PSC1 Disease Severity on Olive Twigs of L. monopetalum Bacterial Endophyte LMRE 36 Towards PSC1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lara, E.; Belbahri, L. SSU rRNA reveals major trends in oomycete evolution. Fungal Divers. 2011, 49, 93–100. [Google Scholar] [CrossRef]
- Olson, A.; Aerts, A.; Asiegbu, F.; Belbahri, L.; Bouzid, O.; Broberg, A.; Canbäck, B.; Coutinho, P.M.; Cullen, D.; Dalman, K.; et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 2012, 194, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Luchi, N.; Ghelardini, L.; Belbahri, L.; Quartier, M.; Santini, A. Rapid detection of Ceratocystis platani inoculum by quantitative Real-Time PCR assay. Appl. Environ. Microbiol. 2013, 79, 5394–5404. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Vercauteren, A.; Heungens, K.; Belbahri, L.; Rigling, D. Phytophthora diversity and the population structure of Phytophthora ramorum in Swiss ornamental nurseries. Plant Pathol. 2013, 62, 1063–1071. [Google Scholar] [CrossRef]
- Abad, Z.G.; Abad, J.A.; Cunnington, J.H.; Smith, I.W.; Blomquist, C.; Balci, Y.; Moralejo, E.; Perez-Sierra, A.; Abad-Campos, P.; Alvarez-Bernaola, L.A.; et al. Phytophthora niederhauserii sp. nov. a new polyphagous species mostly isolated from ornamentals potted plants in twelve countries of five continents. Mycologia 2014, 106, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Alenezi, F.N.; Weitz, H.J.; Belbahri, L.; Nidhal, J.; Luptáková, L.; Jaspars, M.; Woodward, S. Draft Genome Sequence of Aneurinibacillus migulanus NCTC 7096. Genome Announc. 2015, 3, e00234-15. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Weitz, H.J.; Belbahri, L.; Ben Rebah, H.; Luptakova, L.; Jaspars, M.; Woodward, S. Draft genome sequence of Aneurinibacillus migulanus strain Nagano. Genome Announc. 2015, 3, e00232-15. [Google Scholar] [CrossRef]
- Belbahri, L.; Alenezi, F.N.; Luptakova, L.; Rateb, M.E.; Woodward, S. Complete genome sequence of Aneurinibacillus migulanus E1, a gramicidin S and d-phenylalanyl-l-propyl diketopiperazine-deficient mutant. Genome Announc. 2015, 3, e01441-15. [Google Scholar] [CrossRef]
- Cherrad, S.; Charnay, A.; Hernandez, C.; Steva, H.; Belbahri, L.; Vacher, S. Emergence of boscalid-resistant strains of Erysiphe necator in French vineyards. Microbiol. Res. 2018, 216, 79–84. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Fraser, S.; Belka, M.; Dogmuş, T.H.; Heckova, Z.; Oskay, F.; Belbahri, L.; Woodward, S. Biological control of Dothistroma needle blight on pine with Aneurinibacillus migulanus. For. Pathol. 2016, 46, 555–558. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Rekik, I.; Bełka, M.; Ibrahim, A.F.; Luptakova, L.; Jaspars, M.; Woodward, S.; Belbahri, L. Strain-level diversity of secondary metabolism in the biocontrol species Aneurinibacillus migulanus. Microbiol. Res. 2016, 182, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Alenezi, F.N.; Rekik, I.; Chenari Bouket, A.; Luptakova, L.; Weitz, H.J.; Rateb, M.E.; Jaspars, M.; Woodward, S.; Belbahri, L. Increased biological activity of Aneurinibacillus migulanus strains correlates with the production of new gramicidin secondary metabolites. Front. Microbiol. 2017, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; Henson, J.; van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mefteh, F.B.; Daoud, A.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Rateb, M.E.; Kadri, A.; Gharsallah, N.; Belbahri, L. Fungal root microbiome from healthy and brittle leaf diseased date palm trees (Phoenix dactylifera L.) reveals a hidden untapped arsenal of antibacterial and broad spectrum antifungal secondary metabolites. Front. Microbiol. 2017, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Mefteh, F.B.; Daoud, A.; Chenari Bouket, A.; Thissera, B.; Kadri, Y.; Cherif-Silini, H.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Oszako, T.; et al. Date palm trees root-derived endophytes as fungal cell factories for diverse bioactive metabolites. Int. J. Mol. Sci. 2018, 19, 1986. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Mosqueda, M.D.C.; Rocha-Granados, M.D.C.; Glick, B.R.; Santoyo, G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018, 208, 25–31. [Google Scholar] [CrossRef]
- Strobel, G. The emergence of endophytic microbes and their biological promise. J. Fungi 2018, 4, 57. [Google Scholar] [CrossRef]
- Slama, H.; Cherif-Silini, H.; Chenari Bouket, A.; Qader, M.; Silini, A.; Yahiaoui, B.; Alenezi, F.N.; Luptakova, L.; Triki, M.A.; Vallat, A.; et al. Screening for Fusarium antagonistic bacteria from contrasting niches designated the endophyte Bacillus halotolerans as plant warden against Fusarium. Front. Microbiol. 2019, 9, 3236. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, L.S. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Pfaffenbichler, N.; Mitter, B. Microbiome applications from lab to field: Facing complexity. Trends Plant Sci. 2019, 24, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.S.; Tang, K.; Guo, S.X. The plant growth-promoting fungus (PGPF) Alternaria sp. A13 markedly enhances Salvia miltiorrhiza root growth and active ingredient accumulation under greenhouse and field conditions. Int. J. Mol. Sci. 2018, 19, 270. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- He, A.L.; Niu, S.Q.; Zhao, Q.; Li, Y.S.; Gou, J.Y.; Gao, H.J.; Suo, S.Z.; Zhang, J.L. Induced salt tolerance of perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. Int. J. Mol. Sci. 2018, 19, 469. [Google Scholar] [CrossRef] [PubMed]
- Abdennabi, R.; Bardaa, S.; Mehdi, M.; Rateb, M.E.; Raab, A.; Alenezi, F.N.; Sahnoun, Z.; Gharsallah, N.; Belbahri, L. Phoenix dactylifera L. sap enhances wound healing in Wistar rats: Phytochemical and histological assessment. Int. J. Biol. Macromol. 2016, 88, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, Z.; Liu, T.; Wang, Y. Biodiversity, phylogeny, and antifungal functions of endophytic fungi associated with Zanthoxylum bungeanum. Int. J. Mol. Sci. 2016, 17, 1541. [Google Scholar] [CrossRef] [PubMed]
- Daoud, A.; Ben Mefteh, F.; Mnafgui, K.; Turki, M.; Jmal, S.; Ben Amar, R.; Ayadi, F.; El-Feki, A.; Abid, L.; Rateb, M.E.; et al. Cardiopreventive effect of ethanolic extract of date palm pollen against isoproterenol induced myocardial infarction in rats through the inhibition of the angiotensin-converting enzyme. Exp. Toxicol. Pathol. 2017, 69, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-López, A.S.; Pintelon, I.; Stevens, V.; Imperato, V.; Timmermans, J.P.; González-Chávez, C.; Carrillo-González, R.; Van Hamme, J.; Vangronsveld, J.; Thijs, S. Seed endophyte microbiome of Crotalaria pumila unpeeled: Identification of plant-beneficial methylobacteria. Int. J. Mol. Sci. 2018, 19, 291. [Google Scholar] [CrossRef]
- Prieto, P.; Schilirò, E.; Maldonado-González, M.; Valderrama, R.; Barroso-Albarracín, J.B.; Mercado-Blanco, J. Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 2011, 62, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Yadav, K. Exploring the potential of endophytes in agriculture: A minireview. Adv. Plants Agric. Res. 2017, 6, 00221. [Google Scholar] [CrossRef]
- Cambrolle, J.; Mancilla-Leytón, J.M.; Muñoz-Vallés, S.; Figueroa-Luque, E.; Luque, T.; Figueroa, M.E. Evaluation of zinc tolerance and accumulation potential of the coastal shrub Limoniastrum monopetalum (L.) Boiss. Environ. Exp. Bot. 2013, 85, 50–57. [Google Scholar] [CrossRef]
- Cambrolle, J.; Mancilla-Leytón, J.M.; Muñoz-Vallés, S.; Figueroa-Luque, E.; Luque, T.; Figueroa, M.E. Effects of copper sulfate on growth and physiological responses of Limoniastrum monopetalum. Environ. Sci. Pollut. Res. 2013, 20, 8839–8847. [Google Scholar] [CrossRef]
- Manousaki, E.; Galanaki, K.; Papadimitriou, L.; Kalogerakis, N. Metal phytoremediation by the halophyte Limoniastrum monopetalum (L.) Boiss: Two contrasting ecotypes. Int. J. Phytoremediat. 2014, 16, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, N.; Falleh, H.; Jallali, I.; Daly, A.B.; Hajlaoui, H.; Smaoui, A.; Abdelly, C.; Ksouri, R. Variation of phenolic composition and biological activities in Limoniastrum monopetalum L. organs. Acta Physiol. Plant. 2012, 34, 87–96. [Google Scholar] [CrossRef]
- Balan, S.S.; Nethaji, R.; Sankar, S.; Jayalakshmi, S. Production of gelatinase enzyme from Bacillus spp. isolated from the sediment sample of Porto Novo Coastal sites. Asian Pac. J. Trop. Biomed. 2012, 2, 1811–1816. [Google Scholar] [CrossRef]
- Trabelsi, R.; Sellami, H.; Gharbi, Y.; Krid, S.; Cheffi, M.; Kammoun, S.; Dammak, M.; Mseddi, A.; Gdoura, R.; Triki, M.A. Morphological and molecular characterization of Fusarium spp. associated with olive trees dieback in Tunisia. 3 Biotech. 2017, 7, 28. [Google Scholar] [CrossRef]
- Bibi, F.; Strobel, G.A.; Naseer, M.I.; Yasir, M.; Khalaf Al-Ghamdi, A.A.; Azhar, E.I. Microbial flora associated with the halophyte Salsola imbricate and its biotechnical potential. Front. Microbiol. 2018, 9, 65. [Google Scholar] [CrossRef]
- Hortova, B.; Novotny, D.; Erban, T. Physiological characteristics and pathogenicity of eight Neofabraea isolates from apples in Czechia. Eur. J. Hortic. Sci. 2014, 79, 327–334. [Google Scholar]
- Yangui, T.; Sayadi, S.; Dhouib, A. Sensitivity of Pectobacterium carotovorum to hydroxytyrosol-rich extracts and their effect on the development of soft rot in potato tubers during storage. Crop. Prot. 2013, 53, 52–57. [Google Scholar] [CrossRef]
- Romero, J.; Raya, M.C.; Roca, L.F.; Agustí-Brisach, C.; Moral, J.; Trapero, A. Phenotypic, molecular and pathogenic characterization of Phlyctema vagabunda, causal agent of olive leprosy. Plant Pathol. 2018, 67, 277–294. [Google Scholar] [CrossRef]
- Gong, Y.; Bai, J.L.; Yang, H.T.; Zhang, W.D.; Xiong, Y.W.; Ding, P.; Qin, S. Phylogenetic diversity and investigation of plant growth-promoting traits of actinobacteria in coastal salt marsh plant rhizospheres from Jiangsu, China. Syst. Appl. Microbiol. 2018, 41, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Qin, S.; Li, W.J.; Dastager, S.G.; Hozzein, W.N. Actinobacteria in special and extreme habitats: Diversity, function roles, and environmental adaptations. Front. Microbiol. 2016, 7, 1415. [Google Scholar] [CrossRef] [PubMed]
- Spence, C.; Alff, E.; Johnson, C.; Ramos, C.; Donofrio, N.; Sundaresan, V.; Bais, H. Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant Biol. 2014, 14, 130. [Google Scholar] [CrossRef]
- Passari, A.K.; Mishra, V.K.; Saikia, R.; Gupta, V.K.; Singh, B.P. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 2015, 6, 273. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef]
- Khan, N.; Martínez-Hidalgo, P.; Ice, T.A.; Maymon, M.; Humm, E.A.; Nejat, N.; Sanders, E.R.; Kaplan, D.; Hirsch, A.M. Antifungal activity of Bacillus species against Fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 2018, 9, 2363. [Google Scholar] [CrossRef]
- Marag, P.S.; Suman, A. Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol. Res. 2018, 214, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Passari, A.K.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Singh, B.P. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 2016, 193, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi-Zecchin, V.J.; Ikeda, A.C.; Hungria, M.; Adamoski, D.; Kava-Cordeiro, V.; Glienke, C.; Galli-Terasawa, L.V. Identification and characterization of endophytic bacteria from corn (Zea mays L.) roots with biotechnological potential in agriculture. AMB Express 2014, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, X.; Yang, Y.; Heeb, S.; Gao, S.; Chan, K.G.; Camara, M.; Gao, K. Functional identification of the prnABCD operon and its regulation in Serratia plymuthica. Appl. Microbiol. Biotechnol. 2018, 102, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.H.; Gururani, M.A.; Chun, S.C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Hongrittipun, P.; Youpensuk, S.; Rerkasem, B. Screening of nitrogen fixing endophytic bacteria in Oryza sativa L. J. Agric. Sci. 2014, 6, 66. [Google Scholar] [CrossRef]
- Delgado, M.; Mendez, J.; Rodriìguez-Herrera, R.; Aguilar, C.N.; Cruz-Hernaìndez, M.; Balagurusamy, N. Characterization of phosphate solubilizing bacteria isolated from the arid soils of a semi-desert region of north-east Mexico. Biol. Agric. Hortic. 2014, 30, 211–217. [Google Scholar] [CrossRef]
- Wani, P.A.; Khan, M.S. Bacillus species enhance growth parameters of chickpea (Cicer arietinum L.) in chromium stressed soils. Food Chem. Toxicol. 2010, 48, 3262–3267. [Google Scholar] [CrossRef]
- Navarro-Torre, S.; Barcia-Piedras, J.M.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Camacho, M.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant Biol. 2017, 19, 249–256. [Google Scholar] [CrossRef]
- Sorty, A.M.; Meena, K.K.; Choudhary, K.; Bitla, U.M.; Minhas, P.S.; Krishnani, K.K. Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L.) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 2016, 180, 872–882. [Google Scholar] [CrossRef]
- Zhao, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Wu, G.H.; Tian, C.Y. Isolation of endophytic plant growth–promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 2016, 73, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.G.D.; Ramarathnam, R.K.A.S.; Savchuk, S.C. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 2005, 37, 955–964. [Google Scholar] [CrossRef]
- El-Deeb, B.; Fayez, K.; Gherbawy, Y. Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their antimicrobial activities. J. Plant Interact. 2013, 8, 56–64. [Google Scholar] [CrossRef]
- Petersen, L.M.; Tisa, L.S. Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 2013, 59, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Raizada, M.N. Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol. 2016, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, V.; Shrivastava, M.; Ali, S.Z.; Sai Shiva Krishna Prasad, V. Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ. Agric. Sci. 2017, 43, 22–34. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.I.; Kowalski, K.P.; Irizarry, I.; Micci, A.; Soares, M.A.; Bergen, M.S. Disease protection and allelopathic interactions of seed-transmitted endophytic Pseudomonads of invasive seed grass (Phragmites australis). Plant Soil 2018, 422, 195–208. [Google Scholar] [CrossRef]
- Raza, W.; Ling, N.; Zhang, R.; Huang, Q.; Xu, Y.; Shen, Q. Success evaluation of the biological control of Fusarium wilts of cucumber, banana, and tomato since 2000 and future research strategies. Crit. Rev. Biotechnol. 2017, 37, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.N.; Shim, J.; You, Y.; Myung, H.; Bang, K.S.; Cho, M.; Oh, B.T. Characterization of lead resistant endophytic Bacillus sp. MN3-4 and its potential for promoting lead accumulation in metal hyperaccumulator Alnus firma. J. Hazard. Mater. 2012, 199, 314–320. [Google Scholar] [CrossRef]
- Ma, Y.; Oliveira, R.S.; Nai, F.; Rajkumar, M.; Luo, Y.; Rocha, I.; Freitas, H. The hyperaccumulator Sedum plumbizincicola harbors metal-resistant endophytic bacteria that improve its phytoextraction capacity in multi-metal contaminated soil. J. Environ. Manag. 2015, 156, 62–69. [Google Scholar] [CrossRef]
- Gond, S.K.; Bergen, M.; Torres, M.S.; White, J.F. Effect of bacterial endophyte on expression of defense genes in Indian popcorn against Fusarium moniliforme. Symbiosis 2015, 66, 133–140. [Google Scholar] [CrossRef]
- Jeong, M.H.; Lee, Y.S.; Cho, J.Y.; Ahn, Y.S.; Moon, J.H.; Hyun, H.N.; Cha, G.S.; Kim, K.Y. Isolation and characterization of metabolites from Bacillus licheniformis MH48 with antifungal activity against plant pathogens. Microb Pathog. 2017, 110, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Nigris, S.; Baldan, E.; Tondello, A.; Zanella, F.; Vitulo, N.; Favaro, G.; Guidolin, V.; Bordin, N.; Telatin, A.; Barizza, E.; et al. Biocontrol traits of Bacillus licheniformis GL174, a culturable endophyte of Vitis vinifera cv. Glera. BMC Microbiol. 2018, 18, 133. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R.; Menezes, R.C.; Grabe, V.; Li, D.; Baldwin, I.T.; Groten, K. A suite of complementary biocontrol traits allows a native consortium of root-associated bacteria to protect their host plant from a fungal sudden-wilt disease. Mol. Ecol. 2019, 28, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Kejela, T.; Thakkar, V.R.; Thakor, P. Bacillus species (BT42) isolated from Coffea arabica L. rhizosphere antagonizes Colletotrichum gloeosporioides and Fusarium oxysporum and also exhibits multiple plant growth promoting activity. BMC Microbiol. 2016, 16, 277. [Google Scholar] [CrossRef] [PubMed]















© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Slama, H.; Triki, M.A.; Chenari Bouket, A.; Ben Mefteh, F.; Alenezi, F.N.; Luptakova, L.; Cherif-Silini, H.; Vallat, A.; Oszako, T.; Gharsallah, N.; et al. Screening of the High-Rhizosphere Competent Limoniastrum monopetalum’ Culturable Endophyte Microbiota Allows the Recovery of Multifaceted and Versatile Biocontrol Agents. Microorganisms 2019, 7, 249. https://doi.org/10.3390/microorganisms7080249
Ben Slama H, Triki MA, Chenari Bouket A, Ben Mefteh F, Alenezi FN, Luptakova L, Cherif-Silini H, Vallat A, Oszako T, Gharsallah N, et al. Screening of the High-Rhizosphere Competent Limoniastrum monopetalum’ Culturable Endophyte Microbiota Allows the Recovery of Multifaceted and Versatile Biocontrol Agents. Microorganisms. 2019; 7(8):249. https://doi.org/10.3390/microorganisms7080249
Chicago/Turabian StyleBen Slama, Houda, Mohamed Ali Triki, Ali Chenari Bouket, Fedia Ben Mefteh, Faizah N. Alenezi, Lenka Luptakova, Hafsa Cherif-Silini, Armelle Vallat, Tomasz Oszako, Neji Gharsallah, and et al. 2019. "Screening of the High-Rhizosphere Competent Limoniastrum monopetalum’ Culturable Endophyte Microbiota Allows the Recovery of Multifaceted and Versatile Biocontrol Agents" Microorganisms 7, no. 8: 249. https://doi.org/10.3390/microorganisms7080249