The Biochemistry and Evolution of the Dinoflagellate Nucleus
Abstract
:1. Introduction
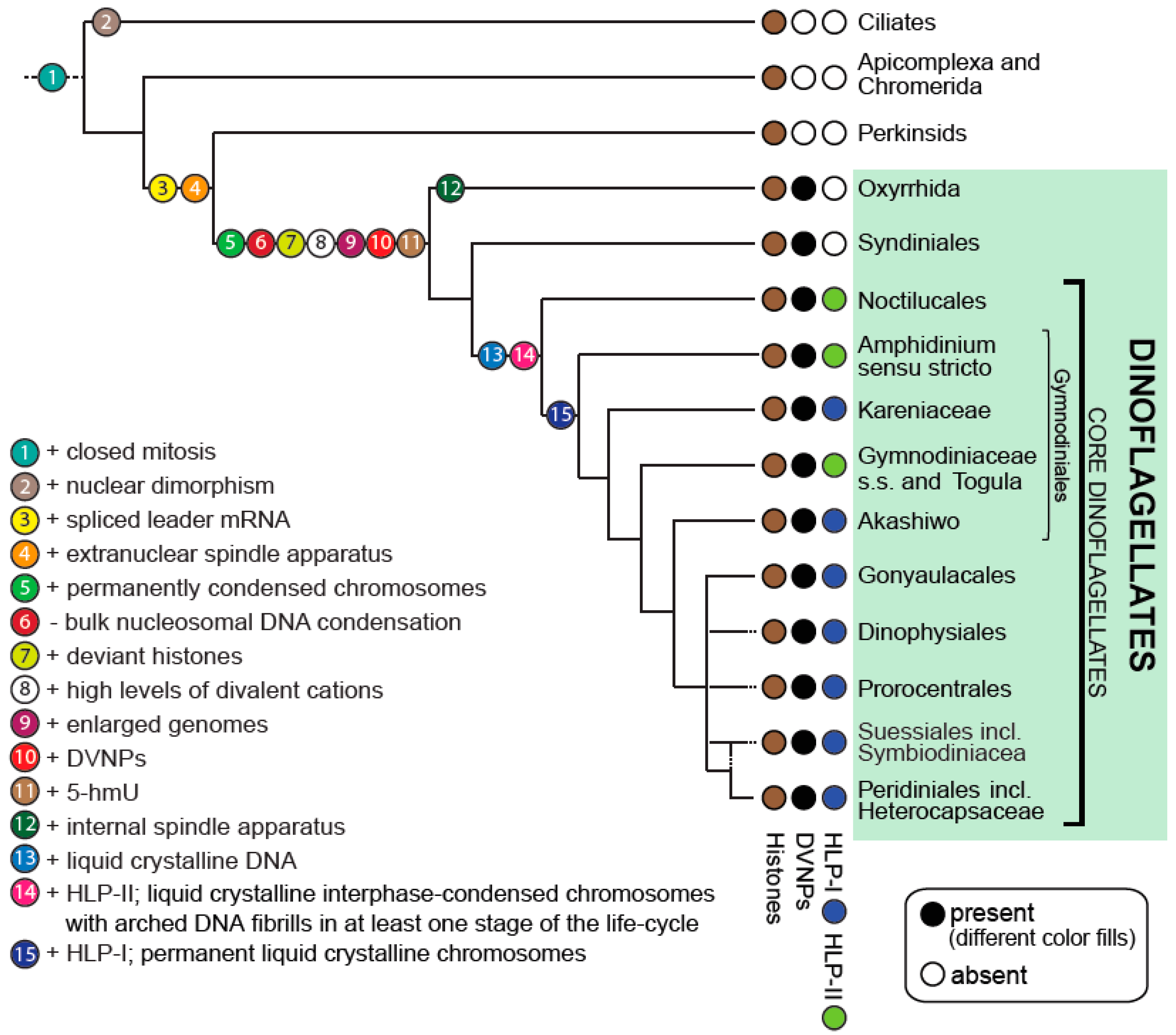
2. Improved Dinoflagellate Phylogenies Provide a Platform for Better Understanding Dinokaryon Development
3. Dinoflagellate Nuclear Biochemistry Is Highly Unusual
3.1. Extremely Large Nuclear DNA Content
3.2. High Concentrations of Bivalent Cations and Transition Metals
3.3. High Amounts of DNA Modifications
3.4. Non-Canonical, Deviant Histones
3.5. Novel Dinoflagellate-Specific Nuclear Proteins
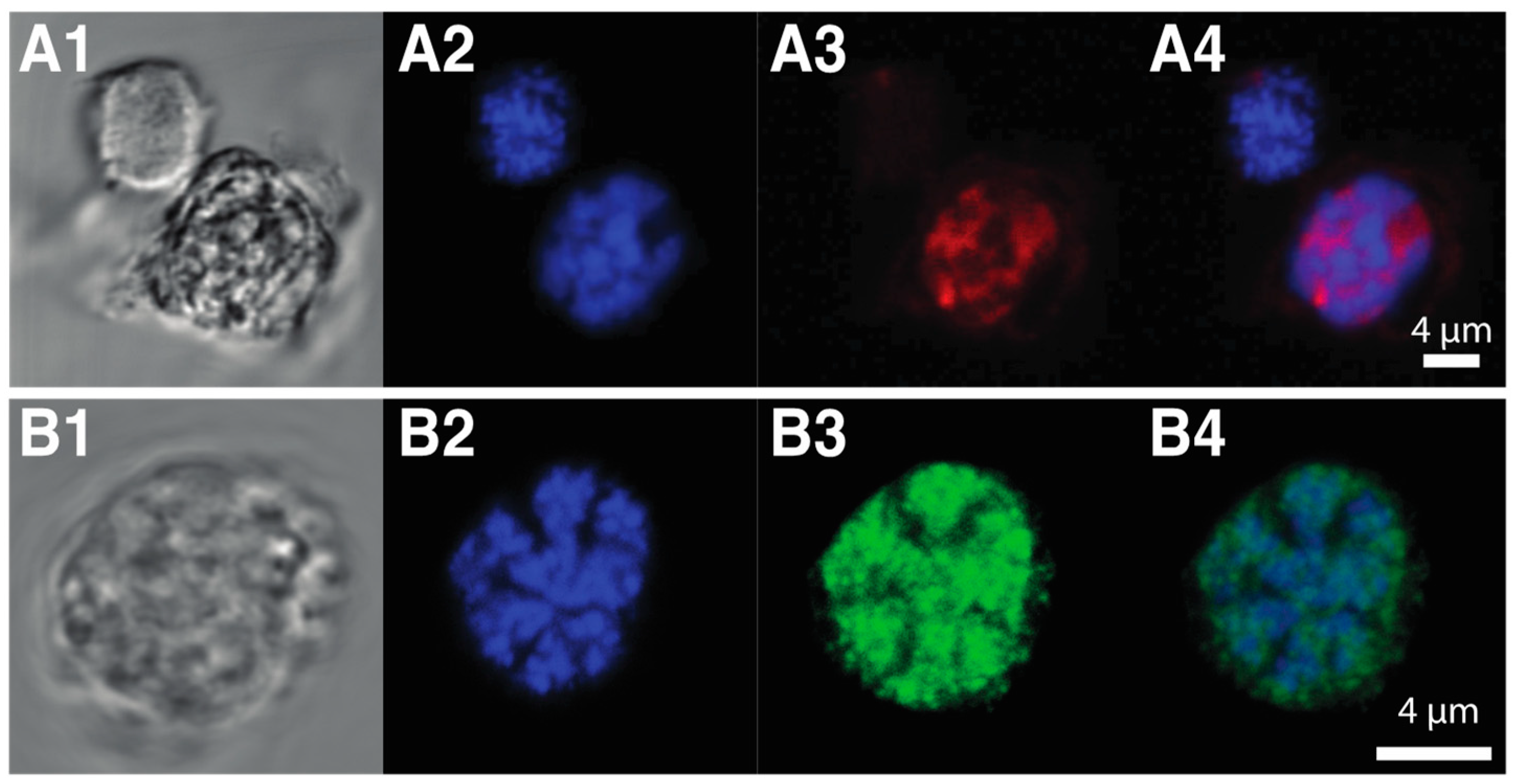
3.5.1. HLPs
3.5.2. DVNPs
4. The Dinoflagellate Chromosome Structure Is Unique amongst Eukaryotes
5. Stepwise Evolution of the Aberrant Dinoflagellate Nucleus
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [PubMed]
- Janouškovec, J.; Gavelis, G.S.; Burki, F.; Dinh, D.; Bachvaroff, T.R.; Gornik, S.G.; Bright, K.J.; Imanian, B.; Strom, S.L.; Delwiche, C.F.; et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA 2017, 114, E171–E180. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Gornik, S.G.; Waller, R.F. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: Character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol. Evol. 2012, 4, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Danne, J.C.; Gornik, S.G.; MacRae, J.I.; McConville, M.J.; Waller, R.F. Alveolate mitochondrial metabolic evolution: Dinoflagellates force reassessment of the role of parasitism as a driver of change in apicomplexans. Mol. Biol. Evol. 2013, 30, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Gómez, F. A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Syst. Biodivers. 2012, 10, 267–275. [Google Scholar] [CrossRef]
- Dodge, J.D. Dinoflagellates: Investigation and phylogenetic speculation. Br. Phycol. J. 1983, 18, 335–356. [Google Scholar] [CrossRef]
- Bachvaroff, T.R.; Handy, S.M.; Place, A.R.; Delwiche, C.F. Alveolate phylogeny inferred using concatenated ribosomal proteins. J. Euk. Microbiol. 2011, 58, 223–233. [Google Scholar] [CrossRef]
- Bachvaroff, T.R.; Gornik, S.G.; Concepcion, G.T.; Waller, R.F.; Mendez, G.S.; Lippmeier, J.C.; Delwiche, C.F. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogenet. Evol. 2014, 70, 314–322. [Google Scholar] [CrossRef]
- Fensome, R.A.; Saldarriaga, J.F.; Taylor, M.F.J.R. Dinoflagellate phylogeny revisited: Reconciling morphological and molecular based phylogenies. Grana 1999, 38, 66–80. [Google Scholar] [CrossRef]
- Saldarriaga, J.F.; Taylor, F.J.; Keeling, P.J.; Cavalier-Smith, T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 2001, 53, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bhattacharya, D.; Lin, S. Phylogeny of dinoflagellates based on mitochondrial cytochrome B and nuclear small subunit rDNA sequence comparisons. J. Phycol. 2005, 41, 411–420. [Google Scholar] [CrossRef]
- Shalchian-Tabrizi, K.; Minge, M.A.; Cavalier-Smith, T.; Nedreklepp, J.M.; Klaveness, D.; Jakobsen, K.S. Combined heat shock protein 90 and ribosomal RNA sequence phylogeny supports multiple replacements of dinoflagellate plastids. J. Euk. Microbiol. 2006, 53, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Hoppenrath, M.; Leander, B.S. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS ONE 2010, 5, e13220-12. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E. Protalveolate phylogeny and systematics and the origins of Sporozoa and dinoflagellates (phylum Myzozoa nom. nov.). Europ. J. Protistol. 2004, 40, 185–212. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Euk. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.; Farmer, M.A.; Andersen, R.A.; Anderson, O.R.; Barta, J.R.; Bowser, S.S.; Brugerolle, G.; Fensome, R.A.; Fredericq, S.; et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Euk. Microbiol. 2005, 52, 399–451. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.; Lane, C.E.; Lukeš, J.; Bass, D.; BowserR, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Euk. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef]
- DeBarry, J.D.; Kissinger, J.C. Jumbled genomes: Missing apicomplexan synteny. Mol. Biol. Evol. 2011, 28, 2855–2871. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Economy, speed and size matter: Evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 2005, 95, 147–175. [Google Scholar] [CrossRef]
- Striepen, B.; Jordan, C.N.; Reiff, S.; van Dooren, G.G. Building the perfect parasite: Cell division in Apicomplexa. PLoS Pathog. 2007, 3, e78. [Google Scholar] [CrossRef] [PubMed]
- Wisecaver, J.H.; Hackett, J.D. Dinoflagellate Genome Evolution. Annu. Rev. Microbiol. 2011, 65, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Jaeckel-Williams, R. Nuclear divisions with reduced numbers of microtubules in Tetrahymena. J. Cell. Sci. 1978, 34, 303–319. [Google Scholar] [PubMed]
- Jenkins, R.A. Fine structure of division in ciliate protozoa. I. Micronuclear mitosis in Blepharisma. J. Cell Biol. 1967, 34, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Azimzadeh, J.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011, 194, 165–175. [Google Scholar] [CrossRef]
- Ausseil, J.; Soyer-Gobillard, M.-O.; Géraud, M.-L.; Bhaud, Y.; Perret, E.; Barbier, M.; Albert, M.; Plaisance, L.; Moreau, H. Dinoflagellate centrosome: Associated proteins old and new. Europ. J. Protistol. 2000, 36, 1–19. [Google Scholar] [CrossRef]
- Triemer, R.E. A Unique mitotic variation in the marine dinoflagellate Oxyrrhis marina (Pyrrophyta). J. Phycol. 1982, 18, 399–411. [Google Scholar] [CrossRef]
- Ris, H.; Kubai, D.F. An unusual mitotic mechanism in the parasitic protozoan Syndinium sp. J. Cell Biol. 1974, 60, 702–720. [Google Scholar] [CrossRef]
- Keeling, P.J.; Slamovits, C.H. Causes and effects of nuclear genome reduction. Curr. Opin. Genet. Dev. 2005, 15, 601–608. [Google Scholar] [CrossRef]
- LaJeunesse, T.C.; Lambert, G.; Andersen, R.A.; Coffroth, M.A.; Galbraith, D.W. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 2005, 41, 880–886. [Google Scholar] [CrossRef]
- Lin, S. The smallest dinoflagellate genome is yet to be found: A comment on LaJeunesse et al. “Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates”. J. Phycol. 2006, 42, 746–748. [Google Scholar] [CrossRef]
- Gornik, S.G.; Ford, K.L.; Mulhern, T.D.; Bacic, A.; McFadden, G.I.; Waller, R.F. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 2012, 22, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, S. Chromosome Evolution in the Genus Ophioglossum. Bot. J. Lin. Soc. 1990, 102, 205–217. [Google Scholar] [CrossRef]
- Zhou, L.; Gui, J.F. Karyotypic diversity in polyploid gibel carp, Carassius auratus gibelio Bloch. Genetica 2002, 115, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Contreras, L.C.; Torres-Mura, J.C.; Spotorno, A.E. The largest known chromosome number for a mammal, in a South American desert rodent. Experientia 1990, 46, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.H.; Lew, A.E.; Jorgensen, W.K.; Barker, S.C. Babesia bovis: Genome size, number of chromosomes and telomeric probe hybridisation. Int. J. Parasitol. 1997, 27, 1569–1573. [Google Scholar] [CrossRef]
- Brayton, K.A.; Lau, A.O.T.; Herndon, D.R.; Hannick, L.; Kappmeyer, L.S.; Berens, S.J.; Bidwell, S.L.; Brown, W.C.; Crabtree, J.; Fadrosh, D.; et al. Genome Sequence of Babesia bovis and Comparative Analysis of Apicomplexan Hemoprotozoa. PLoS Pathog. 2007, 3, e148. [Google Scholar] [CrossRef]
- Leonor Teles-Grilo, M.; Duarte, S.M.; Tato-Costa, J.; Gaspar-Maia, A.; Oliveira, C.; Rocha, A.A.; Marques, A.; Cordeiro-da-Silva, A.; Azevedo, C. Molecular karyotype analysis of Perkinsus atlanticus (Phylum Perkinsozoa) by pulsed field gel electrophoresis. Europ. J. Protistol. 2007, 43, 315–318. [Google Scholar] [CrossRef]
- Marques, A.R.; Tato-Costa, J.; Conde, C.; Azevedo, C.; Teles-Grilo, M.L. Chromosomal localisation of five genes in Perkinsus olseni (Phylum Perkinsozoa). Europ. J. Protistol. 2012, 48, 194–198. [Google Scholar] [CrossRef]
- Herzog, M.; Soyer, M.O. The native structure of dinoflagellate chromosomes and their stabilization by Ca2+ and Mg2+ cations. Europ. J. Cell Biol. 1983, 30, 33–41. [Google Scholar]
- Strick, R.; Strissel, P.L.; Gavrilov, K.; Levi-Setti, R. Cation–chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 2001, 155, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L.; Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010, 2, a000596. [Google Scholar] [CrossRef] [PubMed]
- Levi-Setti, R.; Gavrilov, K.; Rizzo, P. Divalent cation distribution in dinoflagellate chromosomes imaged by high-resolution ion probe mass spectrometry. Europ. J Cell Biol. 2008, 87, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Koltover, I.; Wagner, K.; Safinya, C.R. DNA condensation in two dimensions. Proc. Nat. Acad. Sci. USA 2000, 97, 14046–14051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearns, L.P.; Sigee, D.C. The occurrence of period IV elements in dinoflagellate chromatin: An X-ray microanalytical study. J. Cell. Sci. 1980, 46, 113–127. [Google Scholar] [PubMed]
- Korlach, J.; Turner, S.W. Going beyond five bases in DNA sequencing. Curr. Opin. Struct. Biol. 2012, 22, 251–261. [Google Scholar] [CrossRef]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015, 8, 24. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Gommers-Ampt, J.H.; Borst, P. Hypermodified bases in DNA. FASEB J. 1995, 9, 1034–1042. [Google Scholar] [CrossRef]
- Cadet, J.; Bellon, S.; Douki, T.; Frelon, S.; Gasparutto, D.; Muller, E.; Pouget, J.-P.; Ravanat, J.-L.; Romieu, A.; Sauvaigo, S. Radiation-induced DNA damage: Formation, measurement, and biochemical features. J. Environ. Pathol. Toxicol. Oncol. 2004, 23, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Kallen, R.G.; Simon, M.; Marmur, J. The occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA: 5-hydroxymethyl uracil. J. Mol. Biol. 1962, 5, 248–250. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.R.; Morrissey, L.M.; Foster, L.M.; Geiduschek, E.P. DNA binding by the bacteriophage SPO1-encoded type II DNA-binding protein, transcription factor 1. Formation of nested complexes at a selective binding site. J. Biol. Chem. 1986, 261, 12820–12827. [Google Scholar] [PubMed]
- Rae, P.M.M. 5-Hydroxymethyluracil in the DNA of a dinoflagellate. Proc. Nat. Acad. Sci. USA 1973, 70, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Rae, P. Hydroxymethyluracil in eukaryote DNA: A natural feature of the pyrrophyta (dinoflagellates). Science 1976, 194, 1062–1064. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.; Jakobsen, K.S.; Nordby, O. Characterization of DNA from the dinoflagellate Woloszynskia bostoniensis. J. Protozool. 1988, 35, 418–422. [Google Scholar] [CrossRef]
- Blank, R.J.; Huss, V.A.R.; Kersten, W. Base composition of DNA from symbiotic dinoflagellates: A tool for phylogenetic classification. Arch. Microbiol. 1988, 149, 515–520. [Google Scholar] [CrossRef]
- Steele, R.E.; Rae, P.M.M. Ordered distribution of modified bases in the DNA of a dinoflagellate. Nucleic Acids Res. 1980, 8, 4709–4726. [Google Scholar] [CrossRef] [Green Version]
- Rae, P.M.M.; Steele, R.E. Modified bases in the DNAs of unicellular eukaryotes: An examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems 1978, 10, 37–53. [Google Scholar] [CrossRef]
- De Mendoza, A.; Bonnet, A.; Vargas-Landin, D.B.; Ji, N.; Hong, F.; Yang, F.; Li, L.; Hori, K.; Pflueger, J.; Buckberry, S.; et al. Recurrent acquisition of cytosine methyltransferases into eukaryotic retrotransposons. Nat. Commun. 2018, 9, 1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talbert, P.B.; Henikoff, S. Histone variants—Ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell. Biol. 2010, 11, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R.; Lin, R.; Allis, C.D. Timing of the appearance of ubiquitinated histones in developing new macronuclei of Tetrahymena thermophila. Biochem. Cell Biol. 1991, 69, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Fan, Q.; Cui, L.; Li, J.; Li, J.; Cui, L. The malaria parasite Plasmodium falciparum histones: Organization, expression, and acetylation. Gene 2006, 369, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.J.; Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013, 20, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Talbert, P.B.; Ahmad, K.; Almouzni, G.; Ausió, J.; Berger, F.; Bhalla, P.L.; Bonner, W.M.; Cande, W.; Chadwick, B.P.; Chan, S.W.L.; et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 2012, 5, 7–40. [Google Scholar] [CrossRef]
- Santaguida, S.; Musacchio, A. The life and miracles of kinetochores. EMBO J. 2009, 28, 2511–2531. [Google Scholar] [CrossRef] [Green Version]
- Zlatanova, J.; Thakar, A. H2A.Z: View from the top. Structure 2008, 16, 166–179. [Google Scholar] [CrossRef]
- Petter, M.; Lee, C.C.; Byrne, T.J.; Boysen, K.E.; Volz, J.; Ralph, S.A.; Cowman, A.F.; Brown, G.V.; Duffy, M.F. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011, 7, e1001292-14. [Google Scholar] [CrossRef] [PubMed]
- van Attikum, H.; Gasser, S.M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009, 19, 207–217. [Google Scholar] [CrossRef]
- Ng, R.K.; Gurdon, J.B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2007, 10, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Marzluff, W.F.; Koreski, K.P. Birth and Death of Histone mRNAs. Trends Genet. 2017, 33, 745–759. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Livolant, F.; Bouligand, Y. New observations on the twisted arrangement of dinoflagellate chromosomes. Chromosoma 1978, 68, 21–44. [Google Scholar] [CrossRef]
- Livolant, F.; Bouligand, Y. Double helical arrangement of spread dinoflagellate chromosomes. Chromosoma 1980, 80, 97–118. [Google Scholar] [CrossRef]
- Rizzo, P.J.; Burghardt, R.C. Chromatin structure in the unicellular algae Olisthodiscus luteus, Crypthecodinium cohnii and Peridinium balticum. Chromosoma 1980, 76, 91–99. [Google Scholar] [CrossRef]
- Bodansky, S.; Mintz, L.B.; Holmes, D.S. The mesokaryote Gyrodinium cohnii lacks nucleosomes. Biochem. Biophys. Res. Commun. 1979, 88, 1329–1336. [Google Scholar] [CrossRef]
- Rizzo, P.J. Those amazing dinoflagellate chromosomes. Cell Res. 2003, 13, 215–217. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, P.J. Biochemistry of the dinoflagellate nucleus. In The Biology of Dinoflagellates; Wiley-Blackwell: Hoboken, NJ, USA, 1987; pp. 143–173. [Google Scholar]
- Rizzo, P.J.; Noodén, L.D. Chromosomal Proteins in the Dinoflagellate Alga Gyrodinium cohnii. Science 1972, 176, 796–797. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.J.; Burghardt, R.C. Histone-like protein and chromatin structure in the wall-less dinoflagellate Gymnodinium nelsoni. Biosystems 1982, 15, 27–34. [Google Scholar] [CrossRef]
- Herzog, M.; Soyer, M.O. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species Prorocentrum micans E. Europ. J. Cell Biol. 1981, 23, 295–302. [Google Scholar] [PubMed]
- Kato, K.H.; Moriyama, A.; Huitorel, P.; Cosson, J.; Cachon, M.; Sato, H. Isolation of the major basic nuclear protein and its localization on chromosomes of the dinoflagellate, Oxyrrhis marina. Biol. Cell 1997, 89, 43–52. [Google Scholar] [CrossRef]
- Vernet, G.; Salarovira, M.; Maeder, M.; Jacques, F.; Herzog, M. Basic Nuclear Proteins of the Histone-Less Eukaryote Crypthecodinium cohnii (Pyrrhophyta)—2-dimensional electrophoresis and DNA-binding properties. Biochim. Biophys. Acta 1990, 1048, 281–289. [Google Scholar] [CrossRef]
- Dodge, J.D. Chromosome structure in the dinophyceae. Arch. Microbiol. 1964, 48, 66–80. [Google Scholar] [CrossRef]
- Okamoto, O.K.; Hastings, J.W. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 2003, 321, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.D.; Scheetz, T.E.; Yoon, H.S.; Soares, M.B.; Bonaldo, M.F.; Casavant, T.L.; Bhattacharya, D. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics 2005, 6, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Morse, D. A Full suite of histone and histone modifying genes are transcribed in the dinoflagellate Lingulodinium. PLoS ONE 2012, 7, e34340. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Aranda, M.; Sunagawa, S.; Yum, L.K.; DeSalvo, M.K.; Lindquist, E.; Coffroth, M.A.; Voolstra, C.R.; Medina, M. Symbiodinium transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 2012, 7, e35269. [Google Scholar] [CrossRef]
- Riaz, S.; Sui, Z. Molecular Cloning, Transcriptome profiling, and characterization of histone genes in the dinoflagellate Alexandrium pacificum. J. Microbiol. Biotechnol. 2018, 28, 1185–1198. [Google Scholar] [PubMed]
- Lin, S.; Zhang, H.; Zhuang, Y.; Tran, B.; Gill, J.; Hastings, W. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. USA 2010, 107, 20033–20038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinov, G.K.; Lynch, M. Diversity and divergence of dinoflagellate histone proteins. G3 2016, 6, 397–422. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Liu, Y.; Gorovsky, M.A. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell. Biol. 2006, 26, 7719–7730. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.Y.; New, D.C.; Wong, J.C.W.; Hung, V.K.L. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell 2003, 2, 646–650. [Google Scholar] [CrossRef]
- Sala-Rovira, M.; Geraud, M.L.; Caput, D.; Jacques, F.; Soyer-Gobillard, M.O.; Vernet, G.; Herzog, M. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCc) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta). Chromosoma 1991, 100, 510–518. [Google Scholar] [CrossRef]
- Geraud, M.L.; Salarovira, M.; Herzog, M.; Soyer-Gobillard, M.O. Immunocytochemical localization of the DNA-binding protein HCc during the cell-cycle of the histone-less dinoflagellate Protoctista Crypthecodinium cohnii B. Biol. Cell 1991, 71, 123–134. [Google Scholar] [CrossRef]
- Chan, Y.-H.; Wong, J.T.Y. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acids Res. 2007, 35, 2573–2583. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Liu, M.; Dong, F.; Fan, S.; Yao, Y. A Histone-Like protein induces plasmid DNA to form liquid crystals in vitro and gene compaction in vivo. Int. J. Mol. Sci. 2013, 14, 23842–23857. [Google Scholar] [CrossRef]
- Chudnovsky, Y.; Li, J.F.; Rizzo, P.J.; Hastings, J.W.; Fagan, T.F. Cloning, expression, and characterization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum (Dinophyceae). J. Phycol. 2002, 38, 543–550. [Google Scholar] [CrossRef]
- Taroncher-Oldenburg, G.; Anderson, D.M. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 2000, 66, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Irwin, N.A.T.; Martin, B.J.E.; Young, B.P.; Browne, M.J.G.; Flaus, A.; Loewen, C.J.R.; Keeling, P.J.; Howe, L.J. Viral proteins as a potential driver of histone depletion in dinoflagellates. Nat. Commun. 2018, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Alverca, E.; Franca, S.; Díaz de la Espina, S.M. Topology of splicing and snRNP biogenesis in dinoflagellate nuclei. Biol. Cell 2006, 98, 709–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alverca, E.; Cuadrado, A.; Jouve, N.; Franca, S.; Moreno Díaz de la Espina, S. Telomeric DNA localization on dinoflagellate chromosomes: Structural and evolutionary implications. Cytogenet. Genome Res. 2007, 116, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Fojtová, M.; Wong, J.T.Y.; Dvořáčková, M.; Yan, K.T.H.; Sýkorová, E.; Fajkus, J. Telomere maintenance in liquid crystalline chromosomes of dinoflagellates. Chromosoma 2010, 119, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Endoh, H. New details from the complete life cycle of the red-tide dinoflagellate Noctiluca scintillans (Ehrenberg) McCartney. Europ. J. Protistol. 2006, 42, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, A.; Karpov, S.A.; Guillou, L. The parasitic dinoflagellates Blastodinium spp. inhabiting the gut of marine, planktonic copepods: Morphology, ecology, and unrecognized species diversity. Front. Microbiol. 2012, 3, 305. [Google Scholar] [CrossRef]
- Soyer, M.O. Structure du noyau des Blastodinium (Dinoflagellés parasites). Chromosoma 1971, 33, 70–114. [Google Scholar] [CrossRef] [PubMed]
- Perkins, F.O.; Menzel, R.W. Ultrastructure of sporulation in the oyster pathogen Dermocystidium marinum. J. Invert. Path. 1967, 9, 205–229. [Google Scholar] [CrossRef]
- Bouligand, Y. Twisted fibrous arrangements in biological materials and cholesteric mesophases. Tissue Cell 1972, 4, 189. [Google Scholar] [CrossRef]
- Chow, M.H.; Yan, K.T.H.; Bennett, M.J.; Wong, J.T.Y. Birefringence and DNA condensation of liquid crystalline chromosomes. Eukaryot. Cell 2010, 9, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Mitov, M. Cholesteric liquid crystals in living matter. Soft Matter 2017, 13, 4176–4209. [Google Scholar] [CrossRef] [PubMed]
- Cachon, J.; Sato, H.; Cachon, M.; Sato, Y. Analysis by polarizing microscopy of chromosomal structure among dinoflagellates and its phylogenetic involvement. Biol. Cell 1989, 65, 51–60. [Google Scholar] [CrossRef]
- Oakley, B.R.; Dodge, J.D. Evidence for a double-helically coiled toroidal chromonema in the dinoflagellate chromosome. Chromosoma 1979, 70, 277–291. [Google Scholar] [CrossRef]
- Rill, R.L.; Livolant, F.; Aldrich, H.C.; Davidson, M.W. Electron-microscopy of liquid-crystalline DNA—Direct evidence for cholesteric-like organization of DNA in dinoflagellate chromosomes. Chromosoma 1989, 98, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Triemer, R.E. Chromosome structure and mitosis in the dinoflagellates—An ultrastructural approach to an evolutionary problem. Biosystems 1981, 14, 289–298. [Google Scholar] [CrossRef]
- Grigoryev, S.A.; Woodcock, C.L. Chromatin organization—The 30nm fiber. Exp. Cell Res. 2012, 318, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Horowitz-Scherer, R.A.; Woodcock, C.L. Organization of interphase chromatin. Chromosoma 2005, 115, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Eltsov, M.; MacLellan, K.M.; Maeshima, K.; Frangakis, A.S.; Dubochet, J. Analysis of cryo-electron microscopy images does not support the existence of 30 nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA 2008, 105, 19732–19737. [Google Scholar] [CrossRef]
- Scheffer, M.P.; Eltsov, M.; Frangakis, A.S. Evidence for short-range helical order in the 30 nm chromatin fibers of erythrocyte nuclei. Proc. Natl. Acad. Sci. USA 2011, 108, 16992–16997. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Gavrilov, A.A. Chromatin without the 30 nm fiber: Constrained disorder instead of hierarchical folding. Epigenetics 2014, 9, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Song, Y.; Chen, C.; Shi, J.; Gan, L. Natural chromatin is heterogeneous and self-associates in vitro. Mol. Biol. Cell 2018, 29, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, K.; Hihara, S.; Takata, H. New insight into the mitotic chromosome structure: Irregular folding of nucleosome fibers without 30 nm chromatin structure. Cold Spring Harb. Symp. Quant. Biol. 2011, 75, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L. Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J. Cell. Biol. 1994, 125, 11–19. [Google Scholar] [CrossRef]
- Nishino, Y.; Eltsov, M.; Joti, Y.; Ito, K.; Takata, H.; Takahashi, Y.; Hihara, S.; Frangakis, A.S.; Imamoto, N.; Ishikawa, T.; et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30 nm chromatin structure. EMBO J. 2012, 31, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Joti, Y.; Hikima, T.; Nishino, Y.; Kamda, F.; Hihara, S.; Takata, H.; Ishikawa, T.; Maeshima, K. Chromosomes without a 30 nm chromatin fiber. Nucleus. 2012, 3, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.; Stott, K. Disordered domains in chromatin-binding proteins. Essays Biochem. 2019, 63, 147–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Travers, A. Modelling and DNA topology of compact 2-start and 1-start chromatin fibres. Nucleic Acids Res. 2019, 37, 46. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Yan, S.H.E.N.; Chen, E.J.; Zhai, Z.H. Nuclear assembly of purified Crythecodinium cohnii chromosomes in cell-free extracts of Xenopus laevis eggs. Cell Res. 2000, 10, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Sano, J.; Kato, K.H. Localization and copy number of the protein-coding genes actin, α-tubulin and Hsp90 in the nucleus of a primitive dinoflagellate, Oxyrrhis marina. Zool. Sci. 2009, 26, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.L.; Graves, M.V.; Müller, D.G.; Boland, W.; Delaroque, N. Phycodnaviridae—large DNA algal viruses. Arch. Virol. 2002, 147, 1479–1516. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Colson, P.; Raoult, D.; Koonin, E.V. Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Dunigan, D.D.; Fitzgerald, L.A.; Van Etten, J.L. Phycodnaviruses: A peek at genetic diversity. Virus Res. 2006, 117, 119–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Török, A.; Schiffer, P.H.; Schnitzler, C.E.; Ford, K.; Mullikin, J.C.; Baxevanis, A.D.; Bacic, A.; Frank, U.; Gornik, S.G. The cnidarian Hydractinia echinata employs canonical and highly adapted histones to pack its DNA. Epigenetics Chromatin 2016, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Török, A.; Gornik, S.G. Sperm nuclear basic proteins of marine invertebrates. Results Probl. Cell Differ. 2018, 65, 15–32. [Google Scholar]
- Eagen, K.P. Principles of chromosome architecture revealed by Hi-C. Trends Biochem. Sci. 2018, 43, 469–478. [Google Scholar] [CrossRef]
- Chen, X.; Shen, Y.; Draper, W.; Buenrostro, J.D.; Litzenburger, U.; Cho, S.W.; Satpathy, A.T.; Carter, A.C.; Ghosh, R.P.; East-Seletsky, A.; et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 2016, 13, 1013–1020. [Google Scholar] [CrossRef]
- Furey, T.S. ChIP-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012, 13, 840–852. [Google Scholar] [CrossRef]
- Okamoto, N.; Horák, A.; Keeling, P.J. Description of two species of early branching dinoflagellates, Psammosa pacifica n. g., n. sp. and P. atlantica n. sp. PLoS ONE 2012, 7, e34900. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gornik, S.G.; Hu, I.; Lassadi, I.; Waller, R.F. The Biochemistry and Evolution of the Dinoflagellate Nucleus. Microorganisms 2019, 7, 245. https://doi.org/10.3390/microorganisms7080245
Gornik SG, Hu I, Lassadi I, Waller RF. The Biochemistry and Evolution of the Dinoflagellate Nucleus. Microorganisms. 2019; 7(8):245. https://doi.org/10.3390/microorganisms7080245
Chicago/Turabian StyleGornik, Sebastian G., Ian Hu, Imen Lassadi, and Ross F. Waller. 2019. "The Biochemistry and Evolution of the Dinoflagellate Nucleus" Microorganisms 7, no. 8: 245. https://doi.org/10.3390/microorganisms7080245





