Production and Characterization of an Extracellular Acid Protease from Thermophilic Brevibacillus sp. OA30 Isolated from an Algerian Hot Spring
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Strain OA30
2.2. Screening for Extracellular Proteolytic Activity
2.3. Phenotypic Characterization
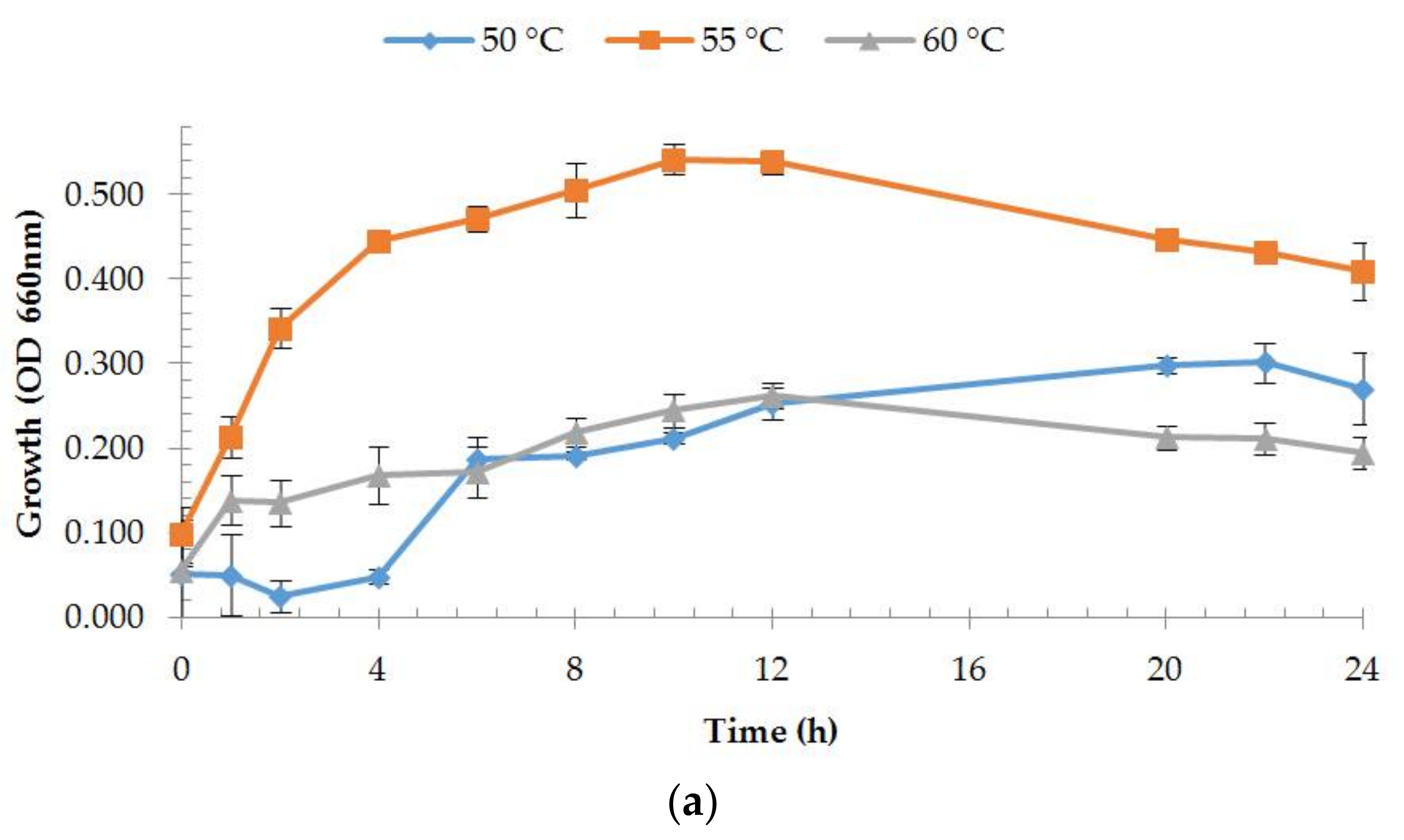
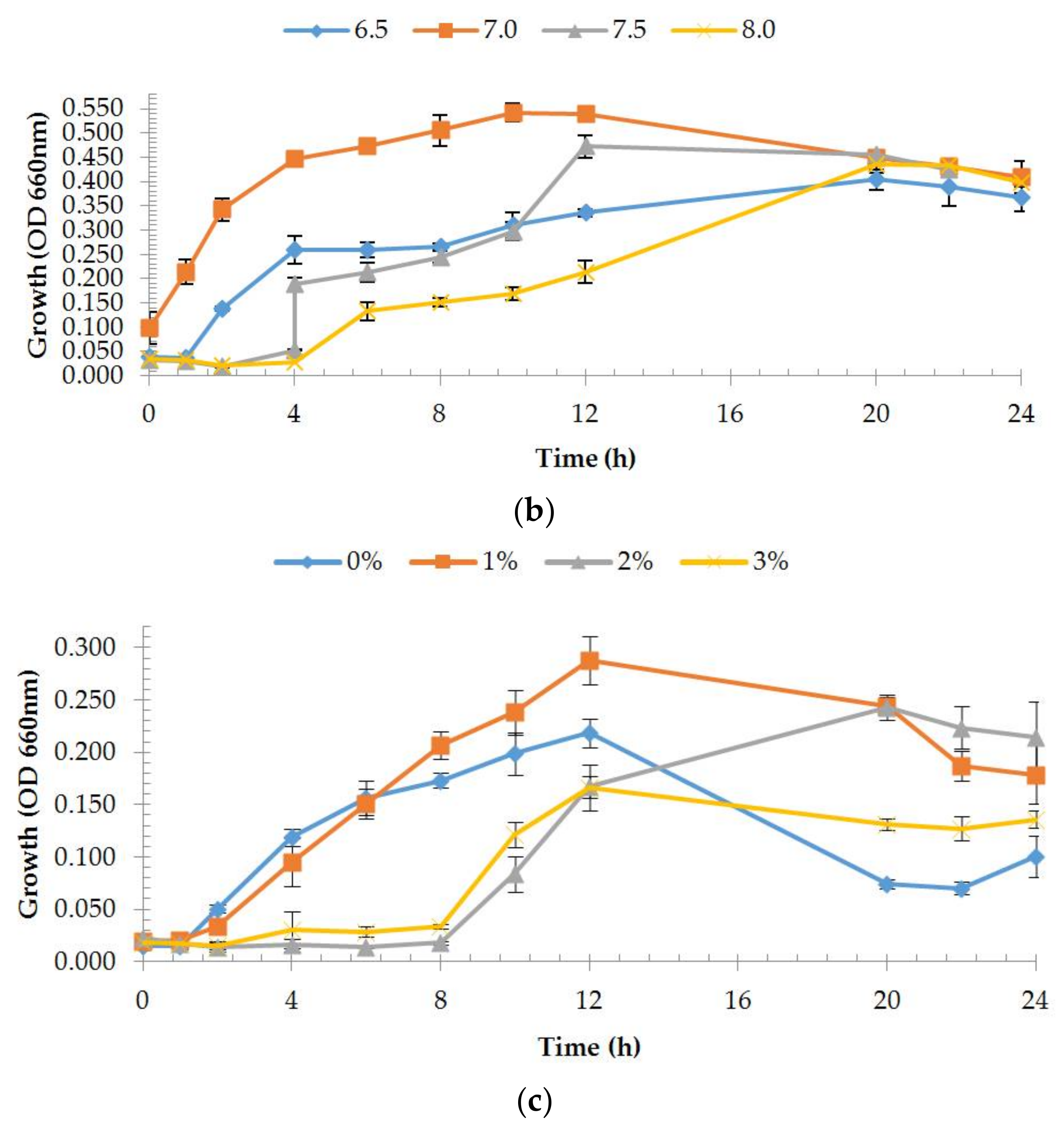
2.4. Estimation of Growth Rates
2.5. Genotypic Characterization
2.5.1. DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
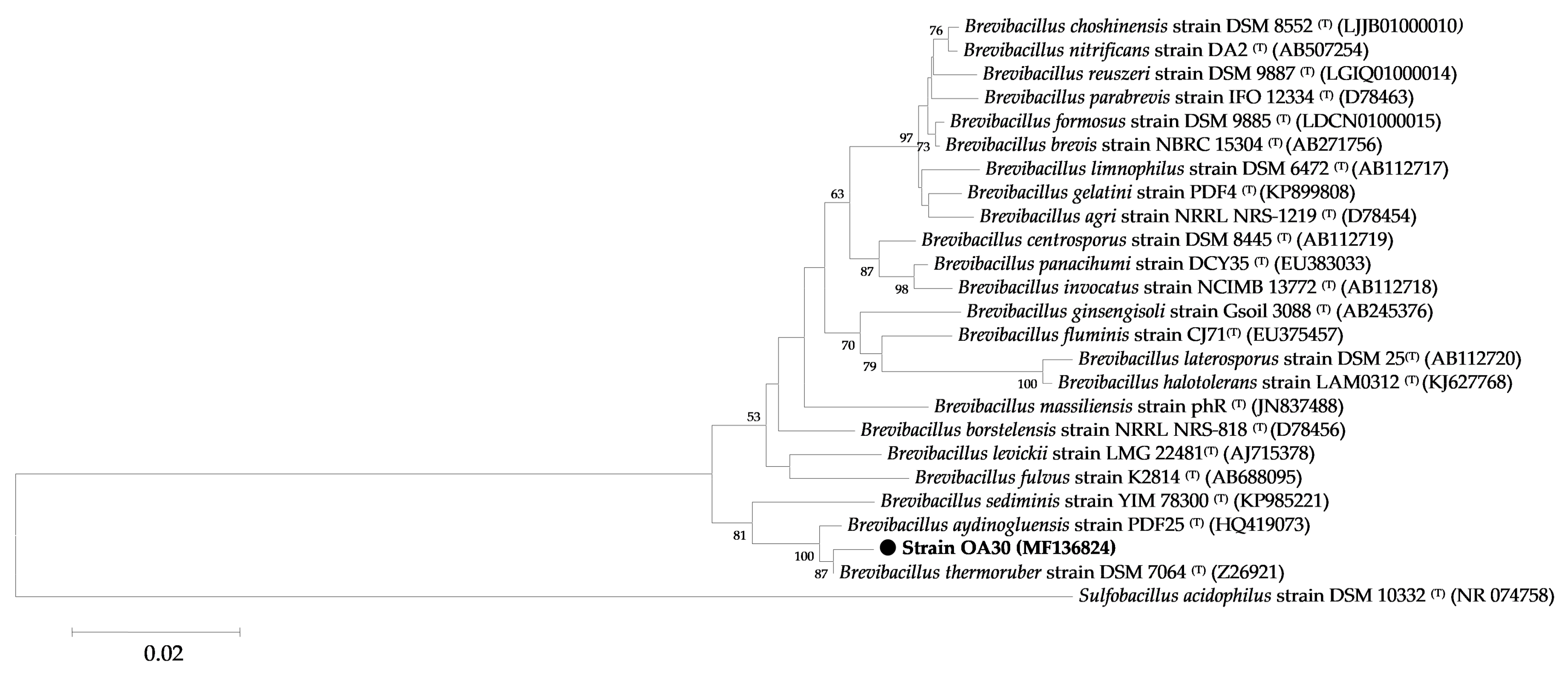
2.5.2. Phylogenetic Analysis
2.6. Enzyme Production
2.7. Purification of Protease
2.8. Molecular Weight Determination and Zymography
2.9. Enzyme Assay
2.10. Effect of Temperature on Protease Activity and Stability
2.11. Effect of pH on Protease Activity
2.12. Effect of Metal Ions on Protease Activity
2.13. Effect of Solvents on Protease Activity
2.14. Effect of Detergents and Chemicals on Protease Activity
2.15. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Characterization of Protease-Producing Strain OA30
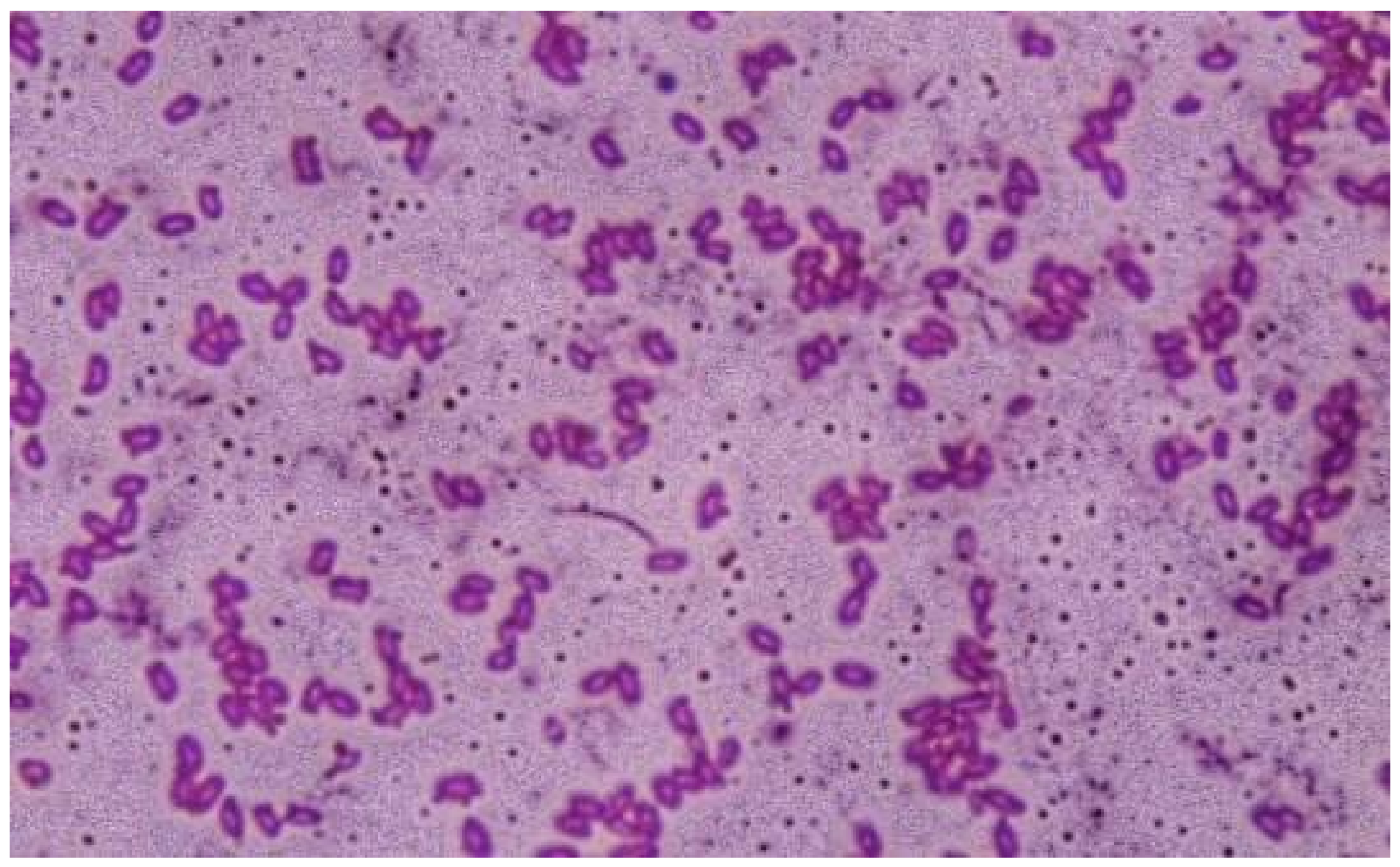
3.1.1. Phenotypic Characterization
3.1.2. Genotypic Characterization
3.2. Protease Purification
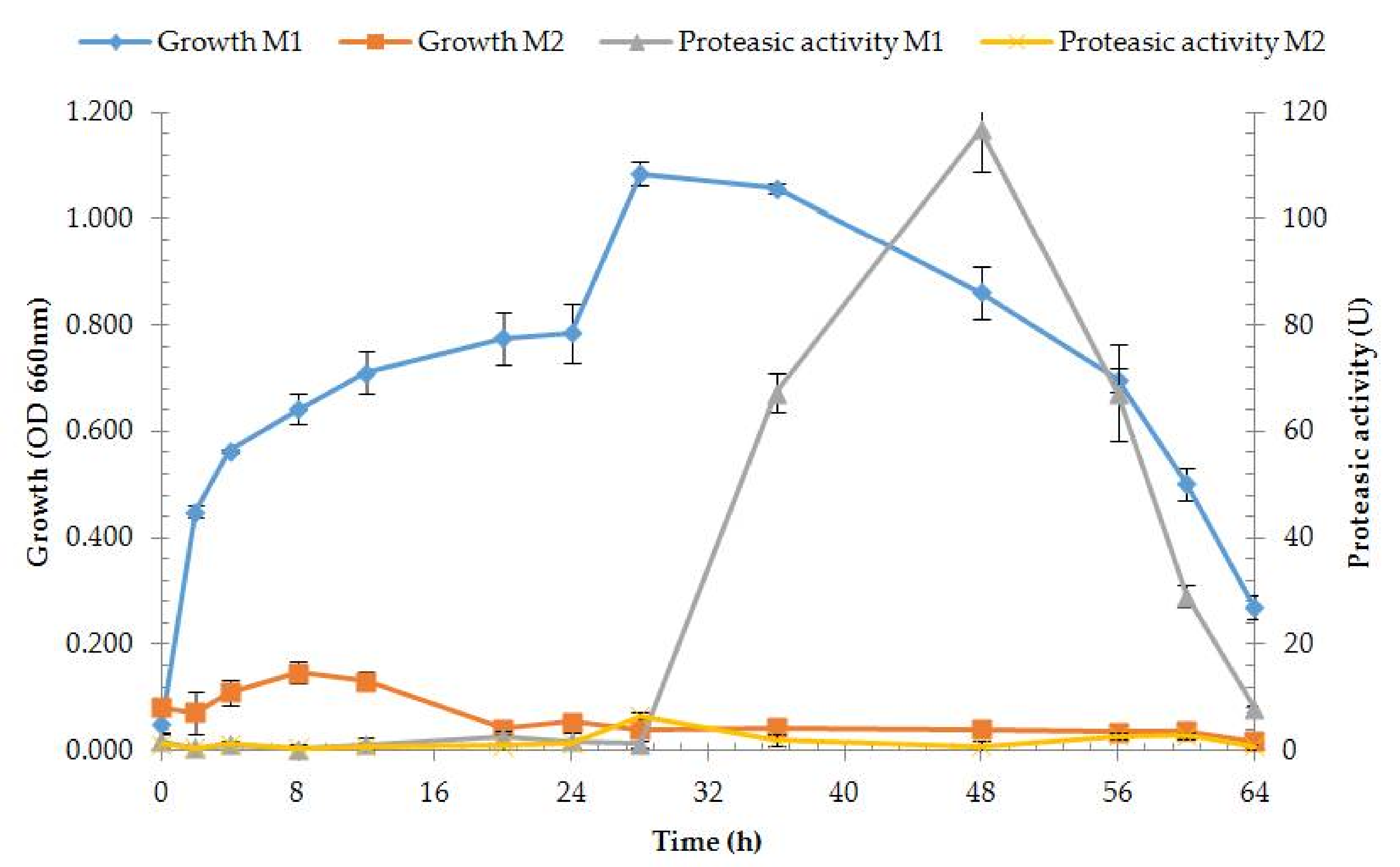
3.2.1. Enzyme Production and Medium Composition Effect
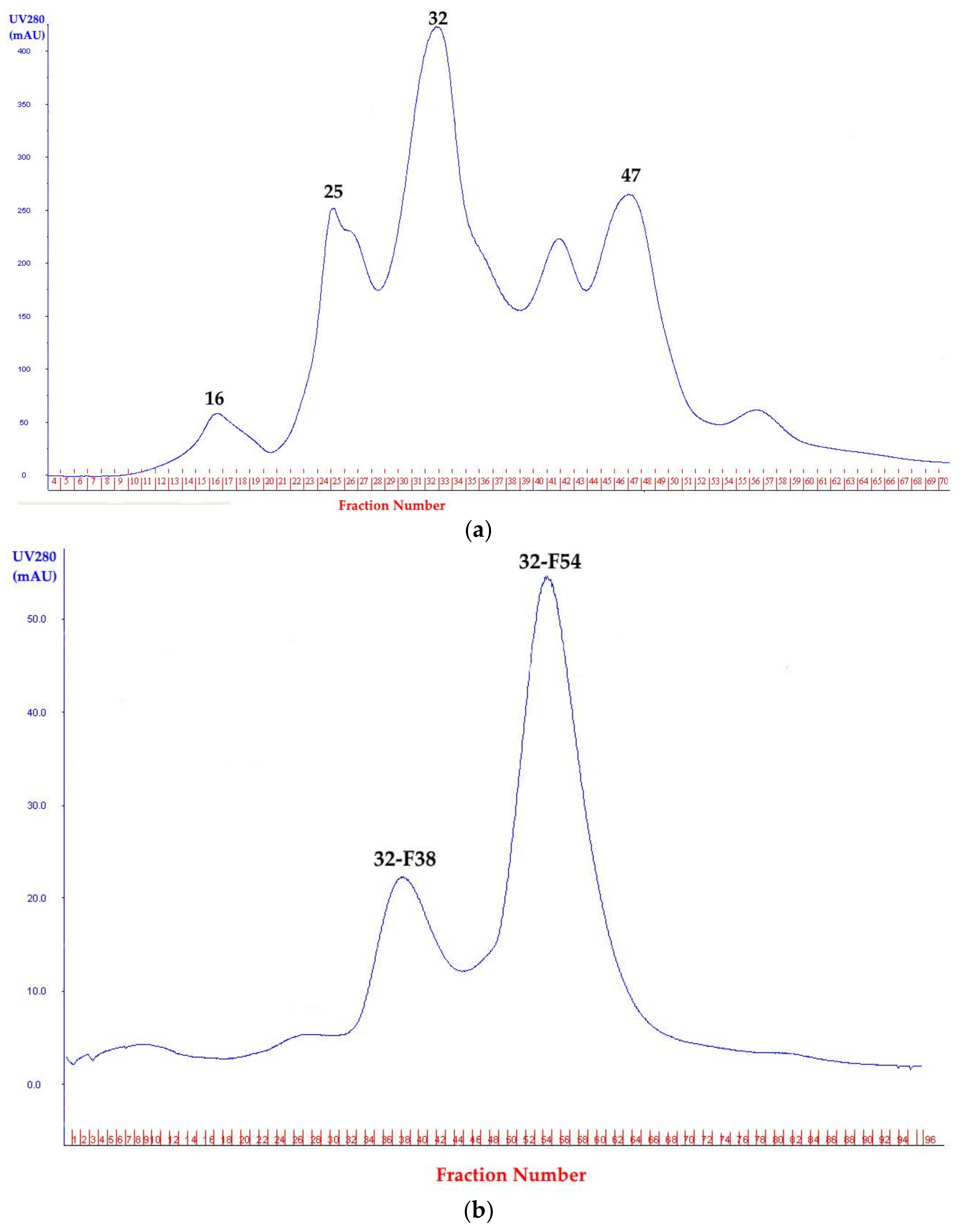
3.2.2. Protease Precipitation, Anion Exchange, and Gel Filtration Chromatography
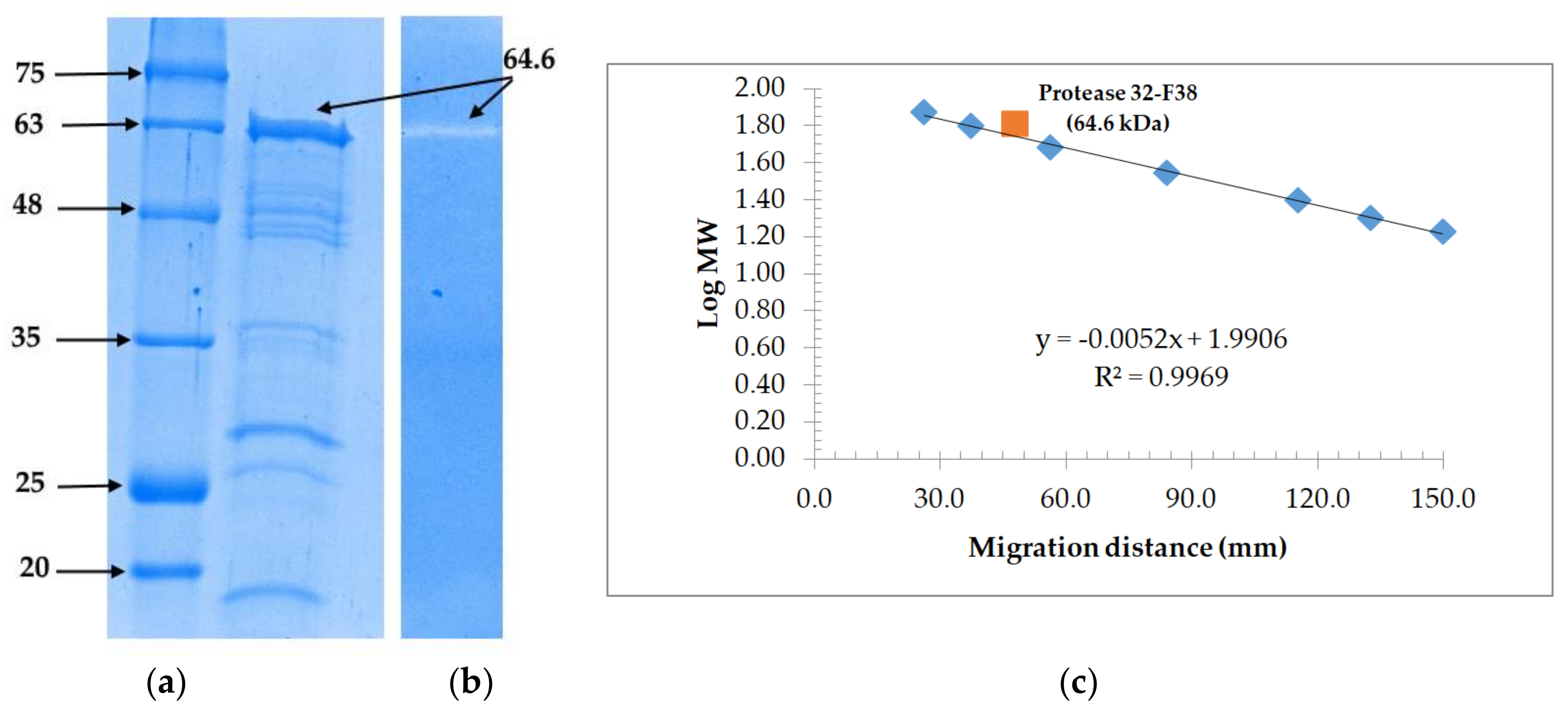
3.3. Molecular Weight Determination and Zymoraphy
3.4. Biochemical Characterization
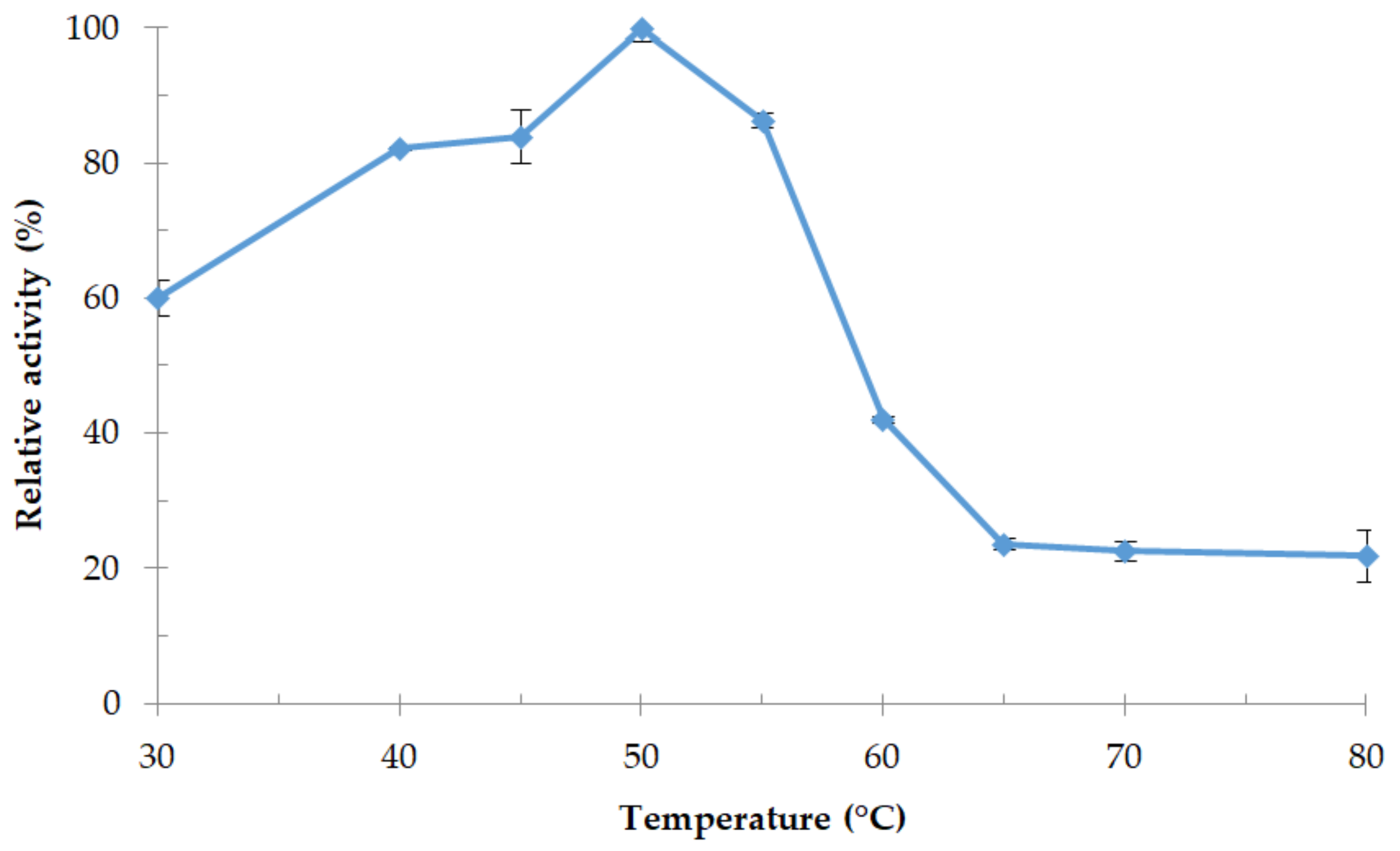
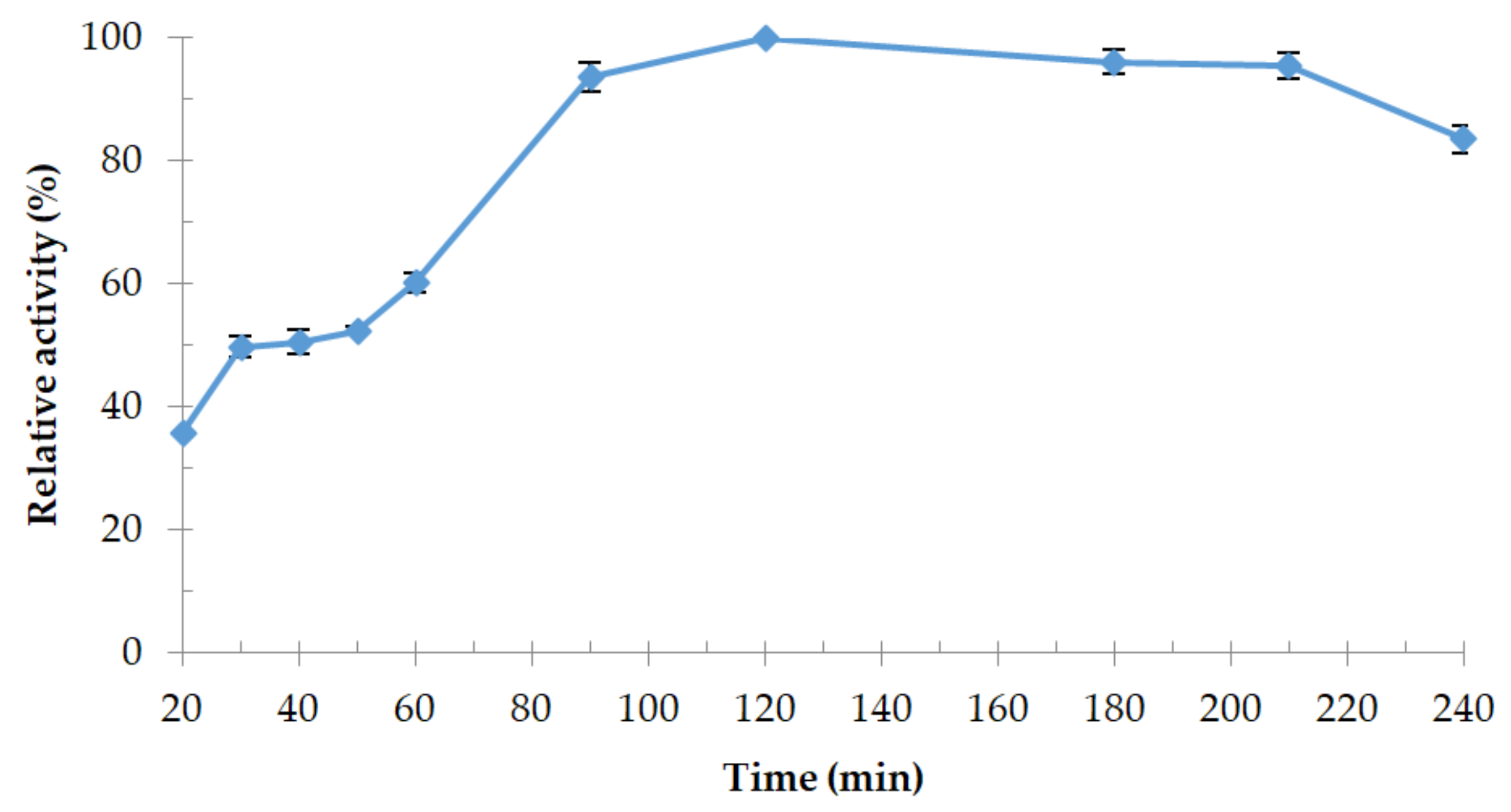
3.4.1. Effect of Temperature and Thermostability of the Enzyme
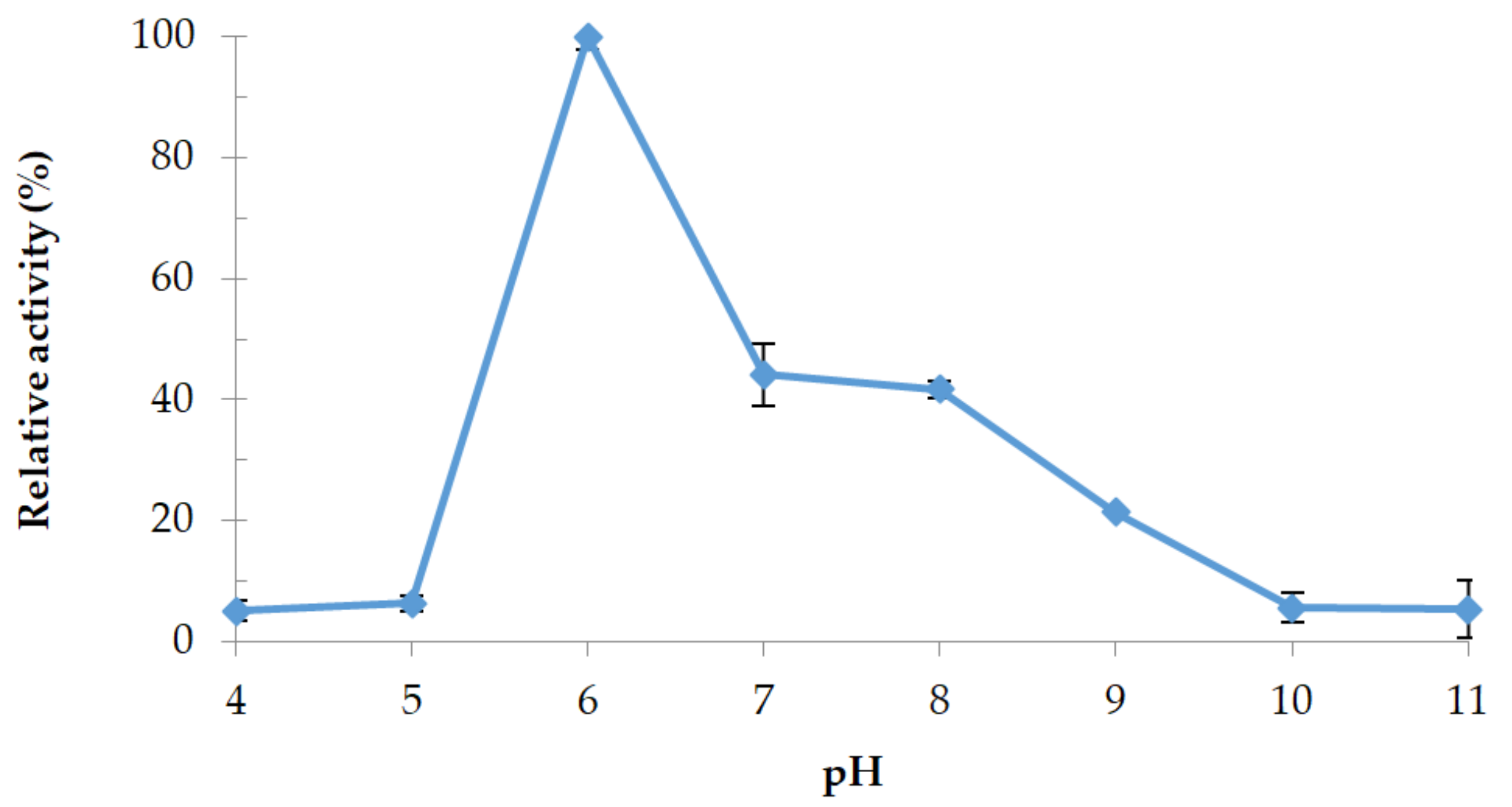
3.4.2. Effect of pH
3.4.3. Effect of Various Chemicals on Protease 32-F38 Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Contesini, F.J.; Melo, R.R.D.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2017, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.M.D.; Bittencourt, M.L.D.A.; Caprara, C.C.; Freitas, M.D.; Almeida, R.P.C.D.; Silveira, D.; Fonseca, Y.M.; Ferreira Filho, E.X.; Pessoa Junior, A.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Chitte, R.; Chaphalkar, S. The World of Proteases Across Microbes, Insects, and Medicinal Trees. In Proteases in Physiology and Pathology; Springer: New York, NY, USA, 2017; pp. 517–526. [Google Scholar]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018. [Google Scholar] [CrossRef]
- Kumar, K.; Arumugam, N.; Permaul, K.; Singh, S. Thermostable enzymes and their industrial applications. Microb. Biotechnol. Interdiscip. Approach 2016, 115–162. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Microbial Enzymes of Use in Industry. In Biotechnology of Microbial Enzymes; Elsevier: New York, NY, USA, 2017; pp. 267–298. [Google Scholar] [CrossRef]
- Feijoo-Siota, L.; Blasco, L.; Rodriguez-Rama, J.; Barros-Velazquez, J.; Miguel, T.; Sanchez-Perez, A.; Villa, T. Recent Patents on Microbial Proteases for the Dairy Industry. Recent Adv. DNA Gene Seq. 2014, 8, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Sawant, R.; Nagendran, S. Protease: An enzyme with multiple industrial applications. World J. Pharm. Sci. 2014, 3, 568–579. [Google Scholar]
- Haki, G.; Rakshit, S. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Mishra, S.S.; Ray, R.C.; Rosell, C.M.; Panda, D. Microbial Enzymes in Food Applications. Microb. Enzyme Technol. Food Appl. 2017, 1. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The genus Geobacillus and their biotechnological potential. In Advances in Applied Microbiology; Elsevier: New York, NY, USA, 2015; Volume 92, pp. 1–48. [Google Scholar]
- Elleuche, S.; Schäfers, C.; Blank, S.; Schröder, C.; Antranikian, G. Exploration of extremophiles for high temperature biotechnological processes. Curr. Opin. Microbiol. 2015, 25, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.D.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B. “Paenibacillaceae” fam. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Manachini, P.; Fortina, M.; Parini, C.; Craveri, R. Bacillus thermoruber sp. nov., nom. rev., a red-pigmented thermophilic bacterium. Int. J. Syst. Evol. Microbiol. 1985, 35, 493–496. [Google Scholar] [CrossRef]
- Allan, R.; Lebbe, L.; Heyrman, J.; De Vos, P.; Buchanan, C.; Logan, N.A. Brevibacillus levickii sp. nov. and Aneurinibacillus terranovensis sp. nov., two novel thermoacidophiles isolated from geothermal soils of northern Victoria Land, Antarctica. Int. J. Syst. Evol. Microbiol. 2005, 55, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.; Morelli, L.; Tompkins, T. Sporeformers as human probiotics: Bacillus, Sporolactobacillus, and Brevibacillus. Compr. Rev. Food Sci. Food Saf. 2003, 2, 101–110. [Google Scholar] [CrossRef]
- Singh, P.K.; Sharma, V.; Patil, P.B.; Korpole, S. Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS ONE 2012, 7, e31498. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.K.; Bisht, S.S.; DeMondal, S.; Kumar, N.S.; Gurusubramanian, G.; Panigrahi, A.K. Brevibacillus as a biological tool: A short review. Antonie Leeuwenhoek 2014, 105, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, S.Y.; Radchenkova, N.; Arga, K.Y.; Kambourova, M.; Oner, E.T. Genomic analysis of Brevibacillus thermoruber 423 reveals its biotechnological and industrial potential. Appl. Microbiol. Biotechnol. 2015, 99, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Media for Environmental Microbiology; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Atlas, R.M. Handbook of Microbiological Media for the Examination of Food; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Logan, N.A.; Berge, O.; Bishop, A.H.; Busse, H.-J.; De Vos, P.; Fritze, D.; Heyndrickx, M.; Kämpfer, P.; Rabinovitch, L.; Salkinoja-Salonen, M.S. Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; Vos, P.D. Brevibacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Bartholomew, J.W.; Mittwer, T. A simplified bacterial spore stain. Stain Technol. 1950, 25, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Frazier, W.C. A method for the detection of changes in gelatin due to bacteria. J. Infect. Dis. 1926. [Google Scholar] [CrossRef]
- Soares, M.M.; Silva, R.d.; Gomes, E. Screening of bacterial strains for pectinolytic activity: Characterization of the polygalacturonase produced by Bacillus sp. Rev. Microbiol. 1999, 30, 299–303. [Google Scholar] [CrossRef]
- Gordon, R.E.; Haynes, W.; Pang, C.H.-N.; Smith, N. The genus Bacillus. In US Department of Agriculture Handbook; Agricultural Research Service: Washington, DC, USA, 1973; pp. 109–126. [Google Scholar]
- Harley, J.P. Laboratory Exercises in Microbiology; McGraw-Hill Science, Engineering & Mathematics: New York, NY, USA, 2004. [Google Scholar]
- Miller, D.; Bryant, J.; Madsen, E.; Ghiorse, W. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [PubMed]
- Farrelly, V.; Rainey, F.A.; Stackebrandt, E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 1995, 61, 2798–2801. [Google Scholar] [PubMed]
- Reysenbach, A.; Pace, N. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction. In Archaea: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 101–107. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2016. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Guendouze, A.; Plener, L.; Bzdrenga, J.; Jacquet, P.; Rémy, B.; Elias, M.; Lavigne, J.-P.; Daudé, D.; Chabrière, E. Effect of Quorum Quenching Lactonase in Clinical Isolates of Pseudomonas aeruginosa and Comparison with Quorum Sensing Inhibitors. Front. Microbiol. 2017, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- StatSoft Inc. Statistica, version 12; Data Analysis Software System; StatSoft Inc.: Tulsa, OK, USA, 2013.
- Bahri, F.; Saibi, H. Characterization, classification, and determination of drinkability of some Algerian thermal waters. Arab. J. Geosci. 2011, 4, 207–219. [Google Scholar] [CrossRef]
- Sinsuwan, S.; Raksakulthai, N.; Rodtong, S.; Yongsawatdigul, J. Production and characterization of proteinases from Brevibacillus sp. isolated from fish sauce fermentation. J. Food Sci. 2006. [Google Scholar] [CrossRef]
- Seswita Zilda, D.; Harmayani, E.N.I.; Widada, J.; Asmara, W.; Irianto, E.K.O.; Patantis, G.; Nuri Fawzya, Y. Optimization of Culture Conditions to Produce Thermostable Keratinolytic Protease of Brevibacillus thermoruber LII, Isolated from the Padang Cermin Hot Spring, Lampung, Indonesia. Microbiol. Indones. 2012, 6, 194–200. [Google Scholar] [CrossRef]
- Wang, S.; Lin, X.; Huang, X.; Zheng, L.; Zilda, D.S. Screening and characterization of the alkaline protease isolated from PLI-1, a strain of Brevibacillus sp. collected from Indonesias hot springs. J. Ocean. Univ. China 2012, 11, 213–218. [Google Scholar] [CrossRef]
- Zilda, D.Z.; Harmayani, E.; Widada, J.; Asmara, W.; Irianto, H.E.; Patantis, G.; Fawzya, Y.N. Purification and characterization of the newly thermostable protease produced by Brevibacillus thermoruber LII isolated From Padang Cermin Hotspring, Indonesia. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Kanekar, P.; Nilegaonkar, S.; Sarnaik, S.; Kelkar, A. Optimization of protease activity of alkaliphilic bacteria isolated from an alkaline lake in India. Bioresour. Technol. 2002, 85, 87–93. [Google Scholar] [CrossRef]
- Esakkiraj, P.; Meleppat, B.; Lakra, A.K.; Ayyanna, R.; Arul, V. Cloning, expression, characterization and application of protease produced by Bacillus cereus PMW8. RSC Adv. 2016, 6, 38611–38616. [Google Scholar] [CrossRef]
- Jaouadi, N.Z.; Rekik, H.; Badis, A.; Trabelsi, S.; Belhoul, M.; Yahiaoui, A.B.; Aicha, H.B.; Toumi, A.; Bejar, S.; Jaouadi, B. Biochemical and molecular characterization of a serine keratinase from Brevibacillus brevis US575 with promising keratin-biodegradation and hide-dehairing activities. PLoS ONE 2013, 8, e76722. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, W.C.A.D.; Martins, M.L.L. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol. 2004, 35, 91–96. [Google Scholar] [CrossRef]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, R. Interaction between casein and sodium dodecyl sulfate. J. Colloid Interface Sci. 2007, 315, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Fusek, M.; Tang, J. Thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. In Structure and Function of the Aspartic Proteinases; Springer: New York, NY, USA, 1991; pp. 255–257. [Google Scholar]
- Bajorath, J.; Hinrichs, W.; Saenger, W. The enzymatic activity of proteinase K is controlled by calcium. FEBS J. 1988, 176, 441–447. [Google Scholar] [CrossRef]
- Salleh, A.B.; Rahman, N.Z.R.A.; Basri, M. New Lipases and Proteases; Nova Publishers: Hauppauge, NY, USA, 2006. [Google Scholar]
- Rawlings, N.D. Protease families, evolution and mechanism of action. In Proteases: Structure and Function; Springer: New York, NY, USA, 2013; pp. 1–36. [Google Scholar]
- Van den Burg, B.; Eijsink, V. Thermolysin and related Bacillus metallopeptidases. In Handbook of Proteolytic Enzymes, 3rd ed.; Elsevier: New York, NY, USA, 2013; pp. 540–553. [Google Scholar]
- Casas, M.P.; González, H.D. Enzyme-Assisted Aqueous Extraction Processes. In Water Extraction of Bioactive Compounds; Elsevier: New York, NY, USA, 2018; pp. 333–368. [Google Scholar]
- Guinane, C.M.; Kent, R.M.; Norberg, S.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Generation of the antimicrobial peptide caseicin A from casein by hydrolysis with thermolysin enzymes. Int. Dairy J. 2015, 49, 1–7. [Google Scholar] [CrossRef]
| Assay | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | |||||||||||
| T (°C) | 50 | 55 | 60 | ||||||||
| pH * | 7.0 | 7.0 | 6.5 | 7.0 | 7.5 | 8.0 | 7.0 | 7.0 | 7.0 | 7.0 | |
| [NaCl] (% w/v) | 0 | 1 | 2 | 3 | 0 | ||||||
| Purification Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Yield (%) | Purification (Fold) |
|---|---|---|---|---|---|
| Cell-free supernatant | 131.78 | 147.2 | 1.12 | 100.00 | 1.00 |
| 80% ammonium sulfate | 32.09 | 130.1 | 4.05 | 88.38 | 3.63 |
| Dialysis | 11.90 | 128.1 | 10.76 | 87.02 | 9.64 |
| AE chromatography | 2.54 | 30.0 | 11.79 | 20.38 | 10.56 |
| Gel filtration | 1.68 | 27.4 | 16.31 | 18.61 | 14.60 |
| Reagent | Concentration | Relative Activity 1 (%) |
|---|---|---|
| Mg2+ | 2.5 mM | 143.24 ± 2.13 |
| Li2+ | 2.5 mM | 100.29 ± 3.25 |
| Fe3+ | 2.5 mM | 0.00 ± 0.00 |
| Cu2+ | 2.5 mM | 7.35 ± 0.00 |
| Zn2+ | 2.5 mM | 0.00 ± 0.00 |
| Mn2+ | 2.5 mM | 113.24 ± 1.85 |
| Ca2+ | 2.5 mM | 90.89 ± 3.45 |
| Ethanol | 1% | 96.46 ± 2.06 |
| Methanol | 1% | 106.19 ± 2.47 |
| Acetone | 1% | 103.54 ± 3.06 |
| SDS | 1% | 438.35 ± 3.56 |
| Tween-20 | 1% | 27.73 ± 2.95 |
| Tween-80 | 1% | 144.54 ± 4.25 |
| Triton X-100 | 1% | 144.25 ± 3.12 |
| EDTA | 1.0 mM | 0.88 ± 0.26 |
| DTT | 1% | 0.36 ± 0.02 |
| PMSF | 1% | 193.59 ± 3.15 |
| Pepstatin A | 1.0 mM | 111.39 ± 3.23 |
| Trypsin inhibitor | 1.0 mM | 186.48 ± 2.23 |
| DMSO | 1% | 87.90 ± 3.56 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomri, M.A.; Rico-Díaz, A.; Escuder-Rodríguez, J.-J.; El Moulouk Khaldi, T.; González-Siso, M.-I.; Kharroub, K. Production and Characterization of an Extracellular Acid Protease from Thermophilic Brevibacillus sp. OA30 Isolated from an Algerian Hot Spring. Microorganisms 2018, 6, 31. https://doi.org/10.3390/microorganisms6020031
Gomri MA, Rico-Díaz A, Escuder-Rodríguez J-J, El Moulouk Khaldi T, González-Siso M-I, Kharroub K. Production and Characterization of an Extracellular Acid Protease from Thermophilic Brevibacillus sp. OA30 Isolated from an Algerian Hot Spring. Microorganisms. 2018; 6(2):31. https://doi.org/10.3390/microorganisms6020031
Chicago/Turabian StyleGomri, Mohamed Amine, Agustín Rico-Díaz, Juan-José Escuder-Rodríguez, Tedj El Moulouk Khaldi, María-Isabel González-Siso, and Karima Kharroub. 2018. "Production and Characterization of an Extracellular Acid Protease from Thermophilic Brevibacillus sp. OA30 Isolated from an Algerian Hot Spring" Microorganisms 6, no. 2: 31. https://doi.org/10.3390/microorganisms6020031







