Chlamydia-Like Organisms (CLOs) in Finnish Ixodes ricinus Ticks and Human Skin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ticks and DNA Extraction
2.2. Skin Samples and DNA Extraction
2.3. DNA Amplification and Sequencing
3. Results
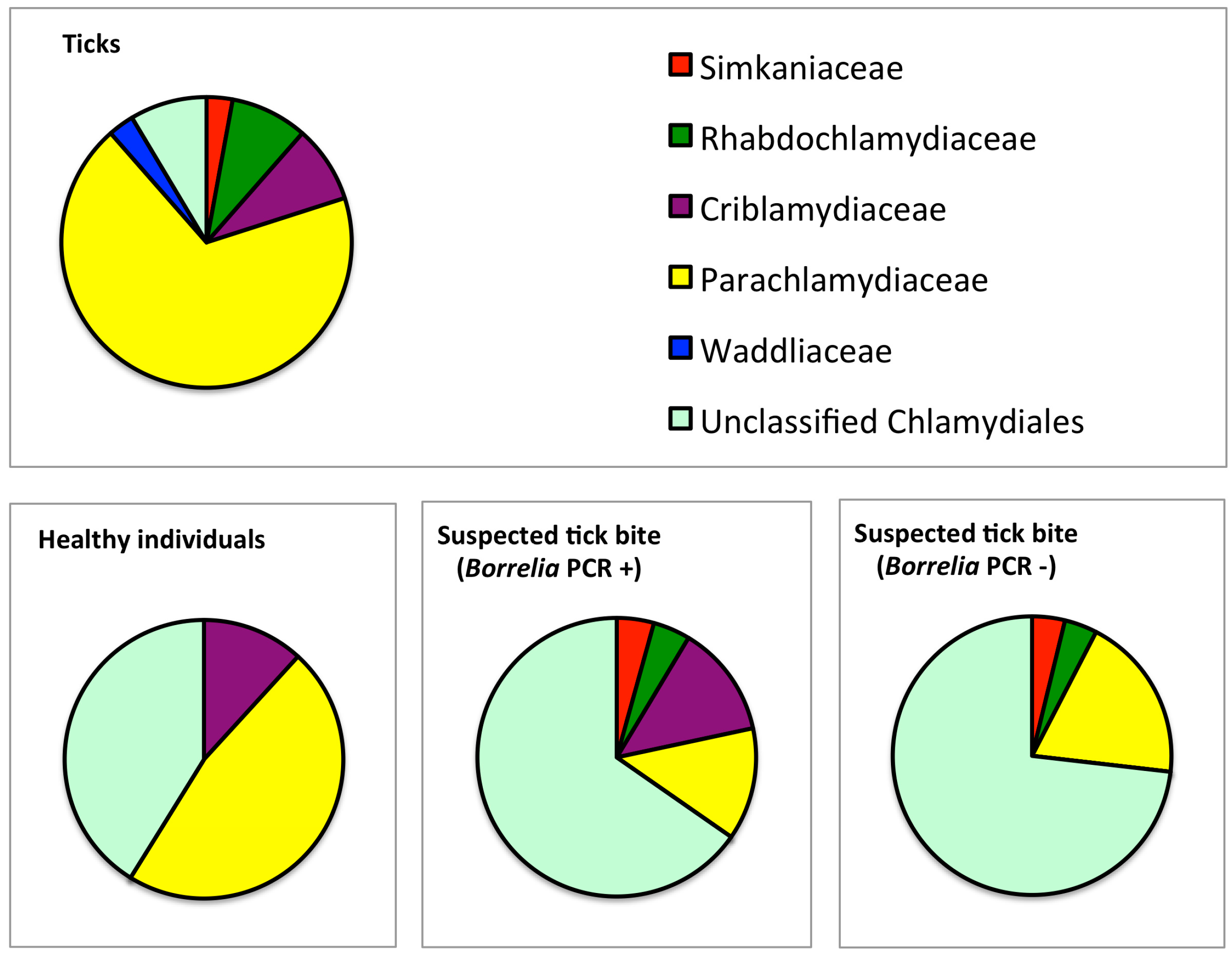
3.1. Prevalence and Sequence Analysis of Chlamydia-Like Organisms (CLOs) in Ticks Collected from Finland
3.2. Prevalence and Sequence Analysis of CLOs in Human Skin
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Everett, K.D.; Bush, R.M.; Andersen, A.A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 1999, 49, 415–440. [Google Scholar] [PubMed]
- Taylor-Brown, A.; Vaughan, L.; Greub, G.; Timms, P.; Polkinghorne, A. Twenty years of research into Chlamydia-like organisms: A revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog. Dis. 2015, 73, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Collingro, A.; Tischler, P.; Weinmaier, T.; Penz, T.; Heinz, E.; Brunham, R.C.; Read, T.D.; Bavoil, P.M.; Sachse, K.; Kahane, S.; et al. Unity in variety—The pan-genome of the Chlamydiae. Mol. Biol. Evol. 2011, 28, 3253–3270. [Google Scholar] [CrossRef] [PubMed]
- Aaziz, R.; Vorimore, F.; Verheyden, H.; Picot, D.; Bertin, C.; Ruettger, A.; Sachse, K.; Laroucau, K. Detection of atypical Chlamydiaceae in roe deer (Capreolus capreolus). Vet. Microbiol. 2015, 181, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Aaziz, R.; Gourlay, P.; Vorimore, F.; Sachse, K.; Siarkou, V.I.; Laroucau, K. Chlamydiaceae in North Atlantic seabirds admitted to a wildlife rescue center in Western France. Appl. Environ. Microbiol. 2015, 81, 4581–4590. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, D.; Venditti, D. Detection of Chlamydiae from freshwater environments by PCR, amoeba coculture and mixed coculture. Res. Microbiol. 2009, 160, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Rieille, N.; Kernif, T.; Bitam, I.; Aeby, S.; Peter, O.; Greub, G. Presence of Chlamydiales DNA in ticks and fleas suggests that ticks are carriers of Chlamydiae. Ticks Tick Borne Dis. 2014, 5, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Brown, A.; Ruegg, S.; Polkinghorne, A.; Borel, N. Characterisation of Chlamydia pneumoniae and other novel Chlamydial infections in captive snakes. Vet. Microbiol. 2015, 178, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Chua, P.K.; Corkill, J.E.; Hooi, P.S.; Cheng, S.C.; Winstanley, C.; Hart, C.A. Isolation of Waddlia malaysiensis, a novel intracellular bacterium, from fruit bat (Eonycteris spelaea). Emerg. Infect. Dis. 2005, 11, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Hokynar, V.E.J.; Lilley, T.M.; Pulliainen, A.T.; Korhonen, S.J.; Paavonen, J.; Puolakkainen, M. Molecular evidence of Chlamydia-like organisms in the faeces of the bat myotis daubentonii. in preparation.
- Verweij, S.P.; Kebbi-Beghdadi, C.; Land, J.A.; Ouburg, S.; Morre, S.A.; Greub, G. Waddlia chondrophila and Chlamydia trachomatis antibodies in screening infertile women for tubal pathology. Microbes. Infect. 2015, 17, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Regan, L.; Greub, G. Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr. Opin. Infect. Dis. 2008, 21, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Pilloux, L.; LeRoy, D.; Brunel, C.; Roger, T.; Greub, G. Mouse model of respiratory tract infection induced by Waddlia chondrophila. PLoS ONE 2016, 11, e0150909. [Google Scholar] [CrossRef] [PubMed]
- Heiskanen-Kosma, T.; Paldanius, M.; Korppi, M. Simkania negevensis may be a true cause of community acquired pneumonia in children. Scand. J. Infect. Dis. 2008, 40, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Jaton, K.; Vaudaux, B.; Greub, G. Parachlamydia and Rhabdochlamydia: Emerging agents of community-acquired respiratory infections in children. Clin. Infect. Dis. 2011, 53, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Niemi, S.; Greub, G.; Puolakkainen, M. Chlamydia-related bacteria in respiratory samples in Finland. Microbes Infect. 2011, 13, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Greub, G.; Boyadjiev, I.; La Scola, B.; Raoult, D.; Martin, C. Serological hint suggesting that Parachlamydiaceae are agents of pneumonia in polytraumatized intensive care patients. Ann. N Y Acad. Sci. 2003, 990, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Greub, G.; Berger, P.; Papazian, L.; Raoult, D. Parachlamydiaceae as rare agents of pneumonia. Emerg. Infect. Dis. 2003, 9, 755–756. [Google Scholar] [CrossRef] [PubMed]
- Greub, G. Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin. Microbiol. Infect. 2009, 15, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Sormunen, J.J.; Penttinen, R.; Klemola, T.; Hanninen, J.; Vuorinen, I.; Laaksonen, M.; Saaksjarvi, I.E.; Ruohomaki, K.; Vesterinen, E.J. Tick-borne bacterial pathogens in Southwestern Finland. Parasit. Vectors. 2016, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Karbowiak, G.; Biernat, B. The role of particular tick developmental stages in the circulation of tick-borne pathogens affecting humans in central europe. 2. Tick-borne encephalitis virus. Ann. Parasitol. 2016, 62, 3–9. [Google Scholar] [PubMed]
- Swanson, S.J.; Neitzel, D.; Reed, K.D.; Belongia, E.A. Coinfections acquired from Ixodes ticks. Clin. Microbiol. Rev. 2006, 19, 708–727. [Google Scholar] [CrossRef] [PubMed]
- Pilloux, L.; Aeby, S.; Gaumann, R.; Burri, C.; Beuret, C.; Greub, G. The high prevalence and diversity of Chlamydiales DNA within Ixodes ricinus ticks suggest a role for ticks as reservoirs and vectors of Chlamydia-related bacteria. Appl. Environ. Microbiol. 2015, 81, 8177–8182. [Google Scholar] [CrossRef] [PubMed]
- Sormunen, J.J.; Klemola, T.; Vesterinen, E.J.; Vuorinen, I.; Hytonen, J.; Hanninen, J.; Ruohomaki, K.; Saaksjarvi, I.E.; Tonteri, E.; Penttinen, R. Assessing the abundance, seasonal questing activity, and Borrelia and tick-borne encephalitis virus (TBEV) prevalence of Ixodes ricinus ticks in a Lyme borreliosis endemic area in Southwest Finland. Ticks Tick Borne Dis. 2016, 7, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Makinen, J.; Vuorinen, I.; Oksi, J.; Peltomaa, M.; He, Q.; Marjamaki, M.; Viljanen, M.K. Prevalence of granulocytic Ehrlichia and Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected from Southwestern Finland and from Vormsi Island in Estonia. APMIS 2003, 111, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Stanek, G.; Wormser, G.P.; Gray, J.; Strle, F. Lyme borreliosis. Lancet 2012, 379, 461–473. [Google Scholar] [CrossRef]
- Ranki, A.; Aavik, E.; Peterson, P.; Schauman, K.; Nurmilaakso, P. Successful amplification of DNA specific for finnish Borrelia burgdorferi isolates in erythema chronicum migrans but not in circumscribed scleroderma lesions. J. Investig. Dermatol. 1994, 102, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, P.; Schroder, M.T.; NiIranen, K.; Nevanlinna, A.; Panelius, J.; Ranki, A. The many faces of solitary and multiple erythema migrans. Acta. Derm. Venereol. 2013, 93, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Norja, P.; Hokynar, K.; Aaltonen, L.M.; Chen, R.; Ranki, A.; Partio, E.K.; Kiviluoto, O.; Davidkin, I.; Leivo, T.; Eis-Hubinger, A.M.; et al. Bioportfolio: Lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc. Natl. Acad. Sci. USA 2006, 103, 7450–7453. [Google Scholar] [CrossRef] [PubMed]
- Hokynar, K.; Soderlund-Venermo, M.; Pesonen, M.; Ranki, A.; Kiviluoto, O.; Partio, E.K.; Hedman, K. A new parvovirus genotype persistent in human skin. Virology 2002, 302, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Lienard, J.; Croxatto, A.; Aeby, S.; Jaton, K.; Posfay-Barbe, K.; Gervaix, A.; Greub, G. Development of a new Chlamydiales-specific real-time PCR and its application to respiratory clinical samples. J. Clin. Microbiol. 2011, 49, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Eddie, B.; Radovsky, F.J.; Stiller, D.; Kumada, N. Psittacosis-lymphogranuloma venereum (PL) agents (Bedsonia, Chlamydia) in ticks, fleas, and native mammals in california. Am. J. Epidemiol. 1969, 90, 449–460. [Google Scholar] [PubMed]
- Facco, F.; Grazi, G.; Bonassi, S.; Magnani, M.; di Pietro, P. Chlamydial and rickettsial transmission through tick bite in children. Lancet 1992, 339, 992–993. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome. Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Park, M.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Temporal stability of the human skin microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Krause, P.J.; Narasimhan, S.; Wormser, G.P.; Rollend, L.; Fikrig, E.; Lepore, T.; Barbour, A.; Fish, D. Human borrelia miyamotoi infection in the united states. N. Engl. J. Med. 2013, 368, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Casson, N.; Greub, G. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture. Environ. Microbiol. 2006, 8, 2125–2135. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Loret, J.F.; Jousset, M.; Greub, G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environ. Microbiol. 2008, 10, 2728–2745. [Google Scholar] [CrossRef] [PubMed]
- Wheelhouse, N.; Sait, M.; Gidlow, J.; Deuchande, R.; Borel, N.; Baily, J.; Caldow, G.; Longbottom, D. Molecular detection of Chlamydia-like organisms in cattle drinking water. Vet. Microbiol. 2011, 152, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Kahane, S.; Greenberg, D.; Newman, N.; Dvoskin, B.; Friedman, M.G. Domestic water supplies as a possible source of infection with simkania. J. Infect. 2007, 54, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Veikkolainen, V.; Vesterinen, E.J.; Lilley, T.M.; Pulliainen, A.T. Bats as reservoir hosts of human bacterial pathogen, bartonella mayotimonensis. Emerg. Infect. Dis. 2014, 20, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Lilley, T.M.; Veikkolainen, V.; Pulliainen, A.T. Molecular detection of candidatus bartonella hemsundetiensis in bats. Vector. Borne Zoonotic. Dis. 2015, 15, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Muller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, C.E.; Turmelle, A.; Kuzmin, I.V. A perspective on lyssavirus emergence and perpetuation. Curr. Opin. Virol. 2011, 1, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Matuschka, F.R.; Fischer, P.; Musgrave, K.; Richter, D.; Spielman, A. Hosts on which nymphal Ixodes ricinus most abundantly feed. Am. J. Trop. Med. Hyg. 1991, 44, 100–107. [Google Scholar] [PubMed]
- Jaenson, T.G.; Jaenson, D.G.; Eisen, L.; Petersson, E.; Lindgren, E. Changes in the geographical distribution and abundance of the tick Ixodes ricinus during the past 30 years in Sweden. Parasit. Vectors 2012, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Pakpour, N.; Yi, E.; Melese, M.; Alemayehu, W.; Bird, M.; Schmidt, G.; Cevallos, V.; Olinger, L.; Chidambaram, J.; et al. Pesky trachoma suspect finally caught. Br. J. Ophthalmol. 2004, 88, 750–751. [Google Scholar] [CrossRef] [PubMed]
- Abrams, J.T.; Vonderheid, E.C.; Kolbe, S.; Appelt, D.M.; Arking, E.J.; Balin, B.J. Sezary T-cell activating factor is a Chlamydia pneumoniae-associated protein. Clin. Diagn. Lab. Immunol. 1999, 6, 895–905. [Google Scholar] [PubMed]
- Rossler, M.J.; Rappl, G.; Muche, M.; Hasselmann, D.O.; Sterry, W.; Tilgen, W.; Reinhold, U. No evidence of skin infection with Chlamydia pneumoniae in patients with cutaneous T cell lymphoma. Clin. Microbiol. Infect. 2003, 9, 721–723. [Google Scholar] [PubMed]
- Carter, J.D.; Gerard, H.C.; Hudson, A.P. Psoriasiform lesions induced by tumour necrosis factor antagonists: A skin-deep medical conundrum. Ann. Rheum. Dis. 2008, 67, 1181–1183. [Google Scholar] [CrossRef] [PubMed]

| Tick Life Stage | No. of Ticks | No. of Positive Specimens 1 | No. of Samples Examined 2 | No. of Positive Samples/Total No. of Individuals (Prevalence of CLO DNA in Individual Ticks %) | No. of Positive Samples/ Total No. of Individuals (Minimum Infection Rate 3) |
|---|---|---|---|---|---|
| Adult | 47 | 19 | 47 | 19/47 (40.4%) | |
| Nymph | 497 | 30 | 215 | 30/497 (6.0%) | |
| Larva | 1282 | 22 | 63 | 22/1282 (1.7%) | |
| Total | 1826 | 71 | 325 | 52/1779 (2.9%) |
| Family-Level (≥90%) 1 | Genus-Level (≥95%) 1 | Species-Level (≥97%) 1 |
|---|---|---|
| Parachlamydiaceae (n = 24) | Parachlamydia (n = 8) | Parachlamydia acanthamoebae (n = 2) |
| Neochlamydia (n = 4) | Neochlamydia sp. | |
| Protochlamydia (n = 1) | Trut23-12-2015_Venoge-Embouchure | |
| Candidatus Metachlamydia (n = 1) | (n = 1) | |
| ND (n = 10) | ND (n = 11) | |
| Rhabdochlamydiaceae (n = 3) | Rhabdochlamydia (n = 3) | Candidatus Rhabdochlamydia porcellionis strain 15C (n = 2) ND (n = 1) |
| Criblamydiaceae (n = 3) | ND (n = 3) | |
| Waddliaceae (n = 1) | ND (n = 1) | |
| Simkaniaceae (n = 1) | ND (n = 1) | |
| Chlamydiaceae | 0 | |
| Unclassified Chlamydiales (n = 3) |
| Family-Level Lineage 1 | Skin Condition (n) | ||
|---|---|---|---|
| Suspected Tick Bite (Borrelia PCR +) (n = 23) | Suspected Tick Bite (Borrelia PCR −) (n = 26) | Healthy Skin (n = 17) | |
| Parachlamydiaceae | 3 | 4 | 8 5 |
| Criblamydiaceae | 3 | 2 6 | |
| Rhabdochlamydiaceae | 1 3 | 2 4 | |
| Simkaniaceae | 1 | 1 | |
| Chlamydiaceae | 0 | 0 | 0 |
| Unclassified Chlamydiales 2 | 15 | 19 | 7 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hokynar, K.; Sormunen, J.J.; Vesterinen, E.J.; Partio, E.K.; Lilley, T.; Timonen, V.; Panelius, J.; Ranki, A.; Puolakkainen, M. Chlamydia-Like Organisms (CLOs) in Finnish Ixodes ricinus Ticks and Human Skin. Microorganisms 2016, 4, 28. https://doi.org/10.3390/microorganisms4030028
Hokynar K, Sormunen JJ, Vesterinen EJ, Partio EK, Lilley T, Timonen V, Panelius J, Ranki A, Puolakkainen M. Chlamydia-Like Organisms (CLOs) in Finnish Ixodes ricinus Ticks and Human Skin. Microorganisms. 2016; 4(3):28. https://doi.org/10.3390/microorganisms4030028
Chicago/Turabian StyleHokynar, Kati, Jani J. Sormunen, Eero J. Vesterinen, Esa K. Partio, Thomas Lilley, Veera Timonen, Jaana Panelius, Annamari Ranki, and Mirja Puolakkainen. 2016. "Chlamydia-Like Organisms (CLOs) in Finnish Ixodes ricinus Ticks and Human Skin" Microorganisms 4, no. 3: 28. https://doi.org/10.3390/microorganisms4030028







