Antibacterial Mechanisms and Antivirulence Activities of Oridonin against Pathogenic Aeromonas hydrophila AS 1.1801
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Chemical Agents
2.2. MIC Determination
2.3. Growth Assay
2.4. Cell Membrane Permeability Assay
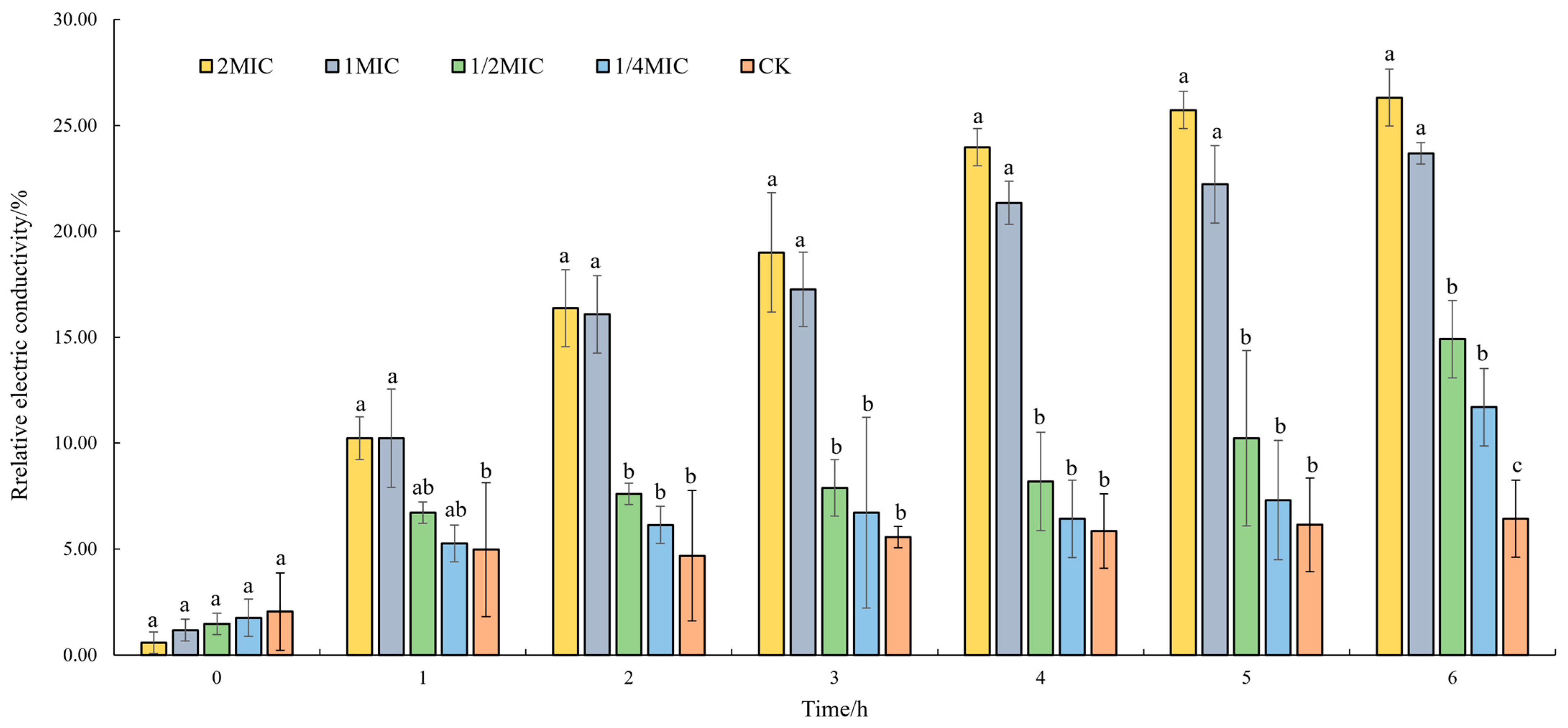
2.4.1. Determination of Conductivity
2.4.2. Determination of Nucleic Acid Release
2.5. Determination of Intracellular Total Protein
2.6. Determination of DNA and RNA Synthesis
2.7. Analysis of Morphological and Physical Changes
2.8. Effect of Oridonin on A. hydrophila Virulence
2.8.1. Lipase Assays
2.8.2. Swarming Assays
2.8.3. Protease Assays
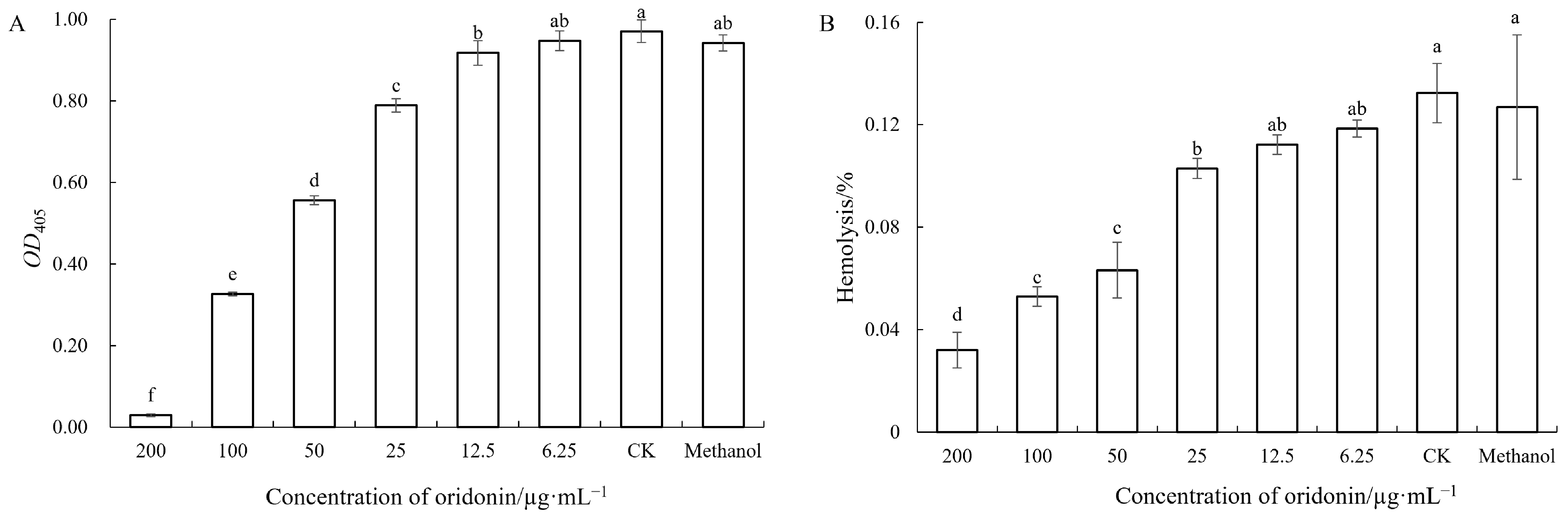
2.8.4. Hemolysis Activity Assays
2.8.5. Antibiofilm Activity Assays
2.9. qRT-PCR
2.10. Statistical Analysis
3. Results
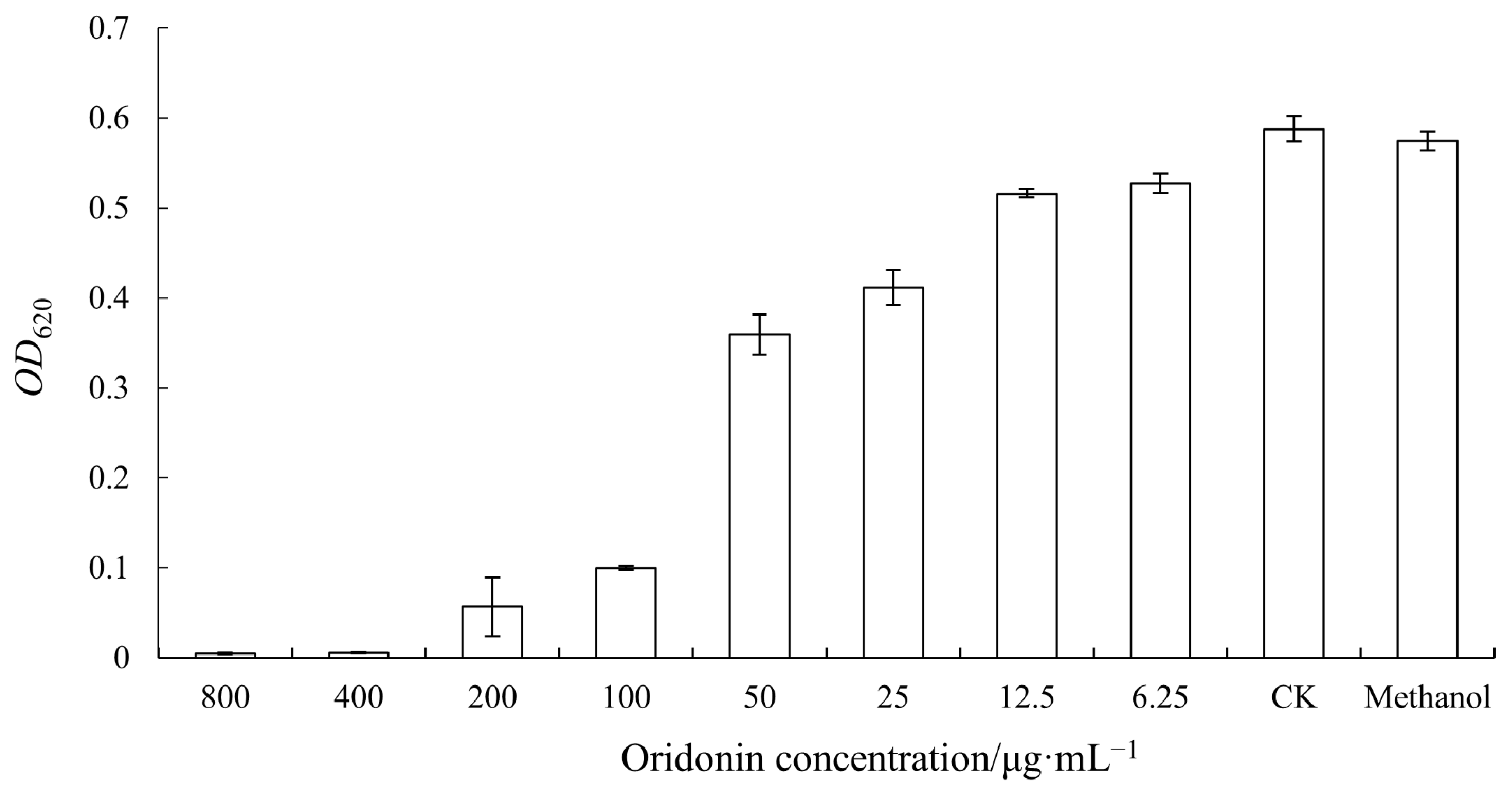
3.1. Determination of MIC of Oridonin against A. hydrophila AS 1.1801
3.2. The Effect of Oridonin on the Growth of A. hydrophila AS 1.1801
3.3. Antibacterial Mechanisms of Oridonin against A. hydrophila AS 1.1801
3.3.1. Effect of Oridonin on the Cell Membrane Permeability
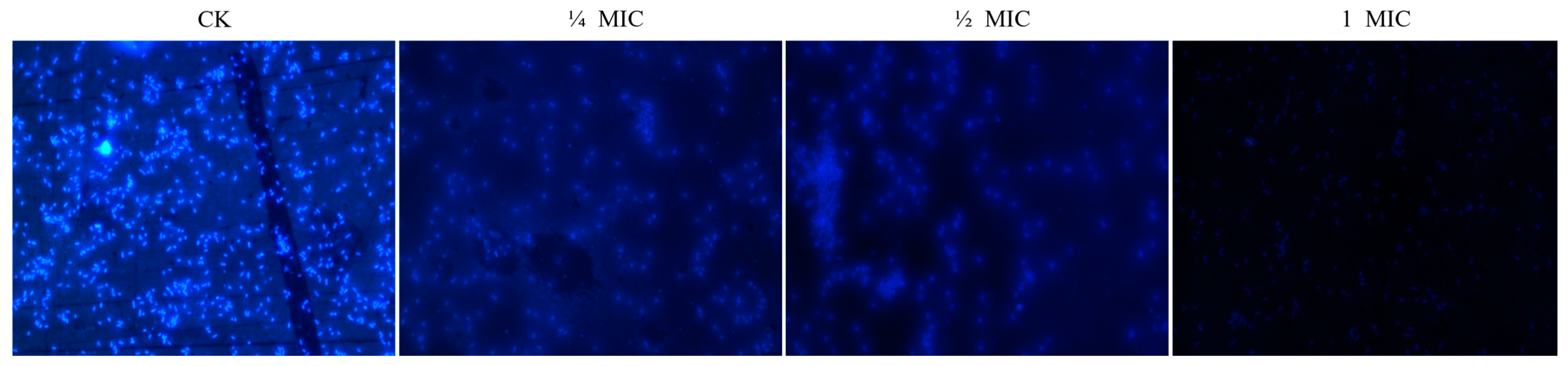
3.3.2. Effect of Oridonin on the Integrity of Cell Membrane
3.4. Effect of Oridonin on the Intracellular Total Protein Synthesis
3.5. Effect of Oridonin on the Synthesis of DNA and RNA
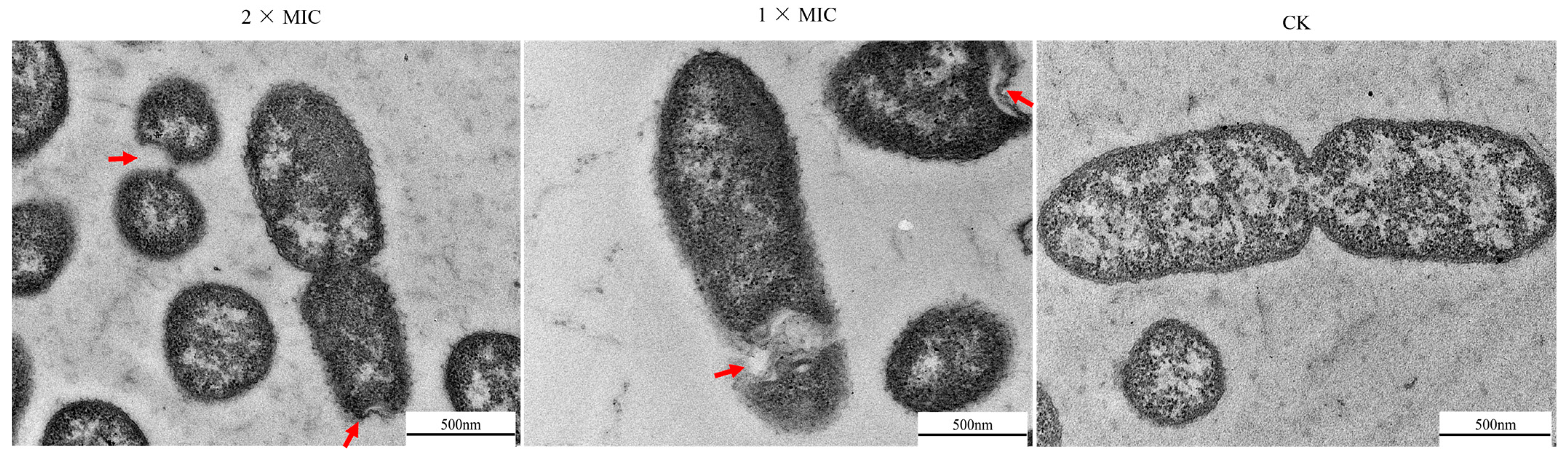
3.6. Effect of Oridonin on the Morphological and Physical Changes of A. hydrophila AS 1.1801
3.7. Inhibitory Effect of Oridonin on the Virulence of A. hydrophila AS 1.1801
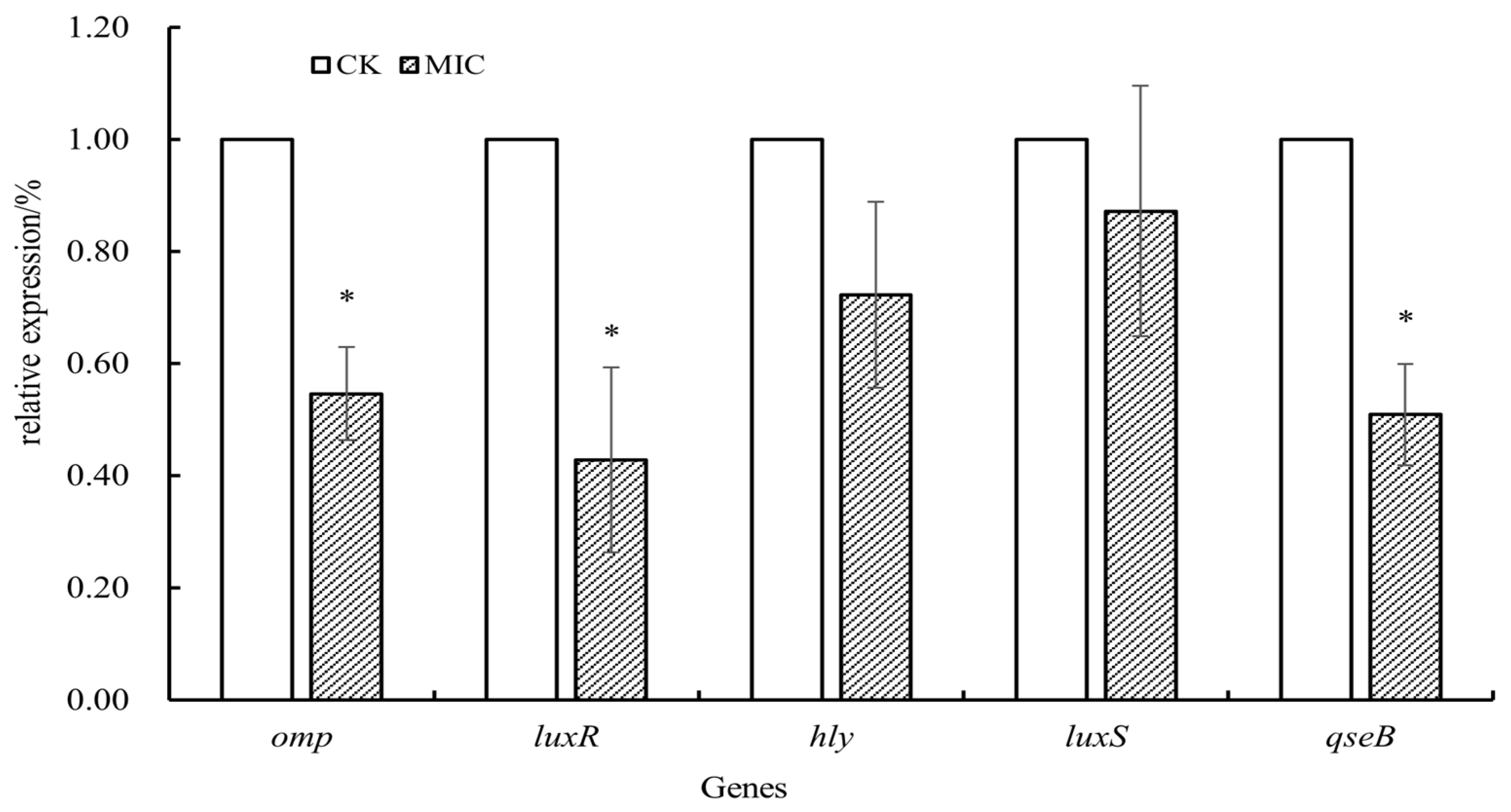
3.8. Effect of Oridonin on the Modulation of A. hydrophila AS 1.1801 Virulence Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, S.; Dahanayake, P.S.; De Silva, B.C.J.; Wickramanayake, M.V.K.S.; Wimalasena, S.H.M.P.; Heo, G.J. Multidrug resistant Aeromonas spp. isolated from zebrafish (Danio rerio): Antibiogram, antimicrobial resistance genes and class 1 integron gene cassettes. Lett. Appl. Microbiol. 2019, 68, 370–377. [Google Scholar] [CrossRef]
- Pu, W.; Guo, G.; Yang, N.; Li, Q.; Yin, F.; Wang, P.; Zheng, J.; Zeng, J. Three species of Aeromonas (A. dhakensis, A. hydrophila and A. jandaei) isolated from freshwater crocodiles (Crocodylus siamensis) with pneumonia and septicemia. Lett. Appl. Microbiol. 2019, 68, 212–218. [Google Scholar] [CrossRef]
- Bilen, S.; Elbeshti, H.T.A.G. A new potential therapeutic remedy against Aeromonas hydrophila infection in rainbow trout (Oncorhynchus mykiss) using tetra, Cotinus coggygria. J. Fish Dis. 2019, 42, 1369–1381. [Google Scholar] [CrossRef]
- De Silva, B.C.J.; Hossain, S.; Dahanayake, P.S.; Heo, G.J. Aeromonas spp. from marketed Yesso scallop (Patinopecten yessoensis): Molecular characterization, phylogenetic analysis, virulence properties and antimicrobial susceptibility. J. Appl. Microbiol. 2019, 126, 288–299. [Google Scholar] [CrossRef]
- Zhao, X.L.; Jin, Z.H.; Di, G.L.; Li, L.; Kong, X.H. Molecular characteristics, pathogenicity and medication regimen of Aeromonas hydrophila isolated from common carp (Cyprinus carpio L.). J. Vet. Med. Sci. 2019, 81, 1769–1775. [Google Scholar] [CrossRef]
- Daskalov, H. The importance of Aeromonas hydrophila in food safety. Food Control 2006, 17, 474–483. [Google Scholar] [CrossRef]
- Chinedu, O.F.; Iniobong, A.D.; Chidinma, W.-E.P. Report on multiple antibiotics resistance Aeromonas hydrophila isolated from catfish farms in Epe Lagos. Middle East J. Appl. Sci. Technol. (MEJAST) 2020, 3, 51–57. Available online: https://www.researchgate.net/publication/345404514 (accessed on 22 September 2020).
- Bardhan, A.; Abraham, T.J. Antibiotic-resistance in motile Aeromonas spp. of Indian major carps sold in retail markets of peri-urban Kolkata, India. J. Aquat. Food Prod. Technol. 2021, 30, 786–793. [Google Scholar] [CrossRef]
- Adah, D.A.; Saidu, L.; Oniye, S.J.; Adah, A.S.; Daoudu, O.B.; Ola-Fadunsin, S.D. Molecular characterization and antibiotics resistance of Aeromonas species isolated from farmed African catfish Clarias gariepinus Burchell, 1822. BMC Vet. Res. 2024, 20, 16. [Google Scholar] [CrossRef]
- Negi, P.S. Plant extracts for the control of bacterial growth: Efficacy, stability and safety issues for food application. Int. J. Food Microbiol. 2012, 156, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.; Coppo, E.; Marchese, A.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Phytochemicals for human disease: An update on plant-derived compounds antibacterial activity. Microbiol. Res. 2017, 196, 44–68. [Google Scholar] [CrossRef]
- Li, S.P.; Zhou, S.; Yang, Q.H.; Liu, Y.T.; Yang, Y.B.; Xu, N.; Ai, X.H.; Dong, J. Cinnamaldehyde decreases the pathogenesis of Aeromonas hydrophila by inhibiting quorum sensing and biofilm formation. Fishes 2023, 8, 122. [Google Scholar] [CrossRef]
- Qin, T.; Chen, K.; Xi, B.W.; Pan, L.K.; Xie, J.; Lu, L.S.; Liu, K. In vitro antibiofilm activity of resveratrol against Aeromonas hydrophila. Antibiotics 2023, 12, 686. [Google Scholar] [CrossRef]
- Lu, L.S.; Wang, J.W.; Qin, T.; Chen, K.; Xie, J.; Xi, B.W. Carvacrol inhibits quorum sensing in opportunistic bacterium Aeromonas hydrophila. Microorganisms 2023, 11, 2027. [Google Scholar] [CrossRef]
- Wang, J.W.; Qin, T.; Chen, K.; Pan, L.K.; Xie, J.; Xi, B.W. Antimicrobial and antivirulence activities of carvacrol against pathogenic Aeromonas hydrophila. Microorganisms 2022, 10, 2170. [Google Scholar] [CrossRef]
- Silva, A.F.E.; Pires, I.C.; da Costa, M.M.; Melo, J.F.B.; Lorenzo, V.P.; de Melo, F.V.S.T.; Copatti, C.E. Antibacterial and antibiofilm activities and synergism with florfenicol from the essential oils of Lippia sidoides and Cymbopogon citratus against Aeromonas hydrophila. J. Appl. Microbiol. 2022, 132, 1802–1812. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.M.; Chen, K.Y.; Yang, L.L.; Tang, T.; Jiang, S.F.; Guo, J.J.; Gao, Z.P. Essential oils from Citrus unshiu Marc. effectively kill Aeromonas hydrophila by destroying cell membrane integrity, influencing cell potential, and leaking intracellular substances. Front. Microbiol. 2022, 13, 869953. [Google Scholar] [CrossRef]
- Hotinger, J.A.; Morris, S.T.; May, A.E. The case against antibiotics and for anti-virulence therapeutics. Microorganisms 2021, 9, 2049. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen-Ivey, C.R.; Figueras, M.J.; McGarey, D.; Liles, M. Virulence factors of Aeromonas hydrophila: In the wake of reclassification. Front. Microbiol. 2016, 7, 1337. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Mizan, M.F.R.; Ha, A.J.; Ha, S.D. Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 2015, 49, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Li, D.H.; Han, T.; Liao, J.; Hu, X.; Xu, S.T.; Tian, K.T.; Gu, X.K.; Cheng, K.G.; Li, Z.L.; Hua, H.M.; et al. Oridonin, a promising ent-Kaurane diterpenoid lead compound. Int. J. Mol. Sci. 2016, 17, 1395. [Google Scholar] [CrossRef]
- Li, D.H.; Han, T.; Xu, S.T.; Zhou, T.T.; Tian, K.T.; Hu, X.; Cheng, K.G.; Li, Z.L.; Hua, H.M.; Xu, J.Y. Antitumor and antibacterial derivatives of oridonin: A main composition of Dong-Ling-Cao. Molecules 2016, 21, 575. [Google Scholar] [CrossRef]
- Yuan, Z.W.; Ouyang, P.; Gu, K.X.; Rehman, T.; Zhang, T.Y.; Yin, Z.Q.; Fu, H.L.; Lin, J.C.; He, C.L.; Shu, G.; et al. The antibacterial mechanism of oridonin against methicillin-resistant Staphylococcus aureus (MRSA). Pharm. Biol. 2019, 57, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Paudel, S.K.; Bhargava, K.; Kotturi, H. Antimicrobial activity of cinnamon oil nanoemulsion against Listeria monocytogenes and Salmonella spp. on melons. LWT-Food Sci. Technol. 2019, 111, 682–687. [Google Scholar] [CrossRef]
- Diao, W.R.; Hu, Q.P.; Zhang, H.; Xu, J.G. Chemical composition, antibacterial activity and mechanism of action of essential oil from seeds of fennel (Foeniculum vulgare Mill.). Food Control 2014, 35, 109–116. [Google Scholar] [CrossRef]
- Kang, J.H.; Song, K.B. Antibacterial activity of the noni fruit extract against Listeria monocytogenes and its applicability as a natural sanitizer for the washing of fresh-cut produce. Food Microbiol. 2019, 84, 103260. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Anh, N.D.Q.; Bossier, P.; Defoirdt, T. Norepinephrine and dopamine increase motility, biofilm formation, and virulence of Vibrio harveyi. Front. Microbiol. 2014, 5, 584. [Google Scholar] [CrossRef]
- Chu, W.H.; Zhou, S.X.; Zhu, W.; Zhuang, X.Y. Quorum quenching bacteria Bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci. Rep. 2014, 4, 5446. [Google Scholar] [CrossRef]
- Luo, G.; Huang, L.X.; Su, Y.Q.; Qin, Y.X.; Xu, X.J.; Zhao, L.M.; Yan, Q.P. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes. Infec. 2016, 5, e85. [Google Scholar] [CrossRef]
- Cui, H.Y.; Zhang, C.H.; Li, C.Z.; Lin, L. Inhibition mechanism of cardamom essential oil on methicillin-resistant Staphylococcus aureus biofilm. LWT-Food Sci. Technol. 2020, 122, 109057. [Google Scholar] [CrossRef]
- Mu, H.B.; Zhang, A.; Zhang, L.; Niu, H.; Duan, J.Y. Inhibitory effects of chitosan in combination with antibiotics on Listeria monocytogenes biofilm. Food Control 2014, 38, 215–220. [Google Scholar] [CrossRef]
- Adékambi, T.; Drancourt, M.; Raoult, D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009, 17, 37–45. [Google Scholar] [CrossRef]
- Tan, H.L.; Chen, K.; Xi, B.W.; Qin, T.; Pan, L.K.; Xie, J. Resveratrol inhibits growth, virulence and biofilm formation of Aeromonas hydrophila. Acta Hydrobiol. Sinica 2019, 43, 861–868. [Google Scholar] [CrossRef]
- Xu, S.T.; Pei, L.L.; Li, D.H.; Yao, H.; Cai, H.; Yao, H.Q.; Wu, X.M.; Xu, J.Y. Synthesis and antimycobacterial evaluation of natural oridonin and its enmein-type derivatives. Fitoterapia 2014, 99, 300–306. [Google Scholar] [CrossRef]
- Li, D.H.; Hu, X.; Han, T.; Xu, S.T.; Zhou, T.T.; Wang, Z.Z.; Cheng, K.G.; Li, Z.L.; Hua, H.M.; Xiao, W.; et al. Synthesis, biological activity, and apoptotic properties of NO-donor/enmein-type ent-kauranoid hybrids. Int. J. Mol. Sci. 2016, 17, 747. [Google Scholar] [CrossRef]
- Meira, T.M.; da Costa, M.M.; Gouveia, J.J.D.; Soares, R.A.N.; Tavares, M.R.S.; Fernandes, A.W.C.; Gouveia, G.V. Action of crude ethanol extract of Hymenaea martiana leaf, gallic acid, and polypyrrole (PPy) against Aeromonas hydrophila. Braz. J. Microbiol. 2023, 54, 1191–1202. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.W.; Wong, G.C. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr. Opin. Solid State Mater. Sci. 2013, 17, 151–163. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Wu, Q.P.; Zhang, J.M.; Yang, X.H. Effects of ozone on membrane permeability and ultrastructure in Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 111, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Heuzenroeder, M.W.; Wong, C.Y.; Flower, R.L. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp. correlation with virulence in a suckling mouse model. FEMS Microbiol. Lett. 1999, 174, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Song, T.Y.; Toma, C.; Nakasone, N.; Iwanaga, M. Aerolysin is activated by metalloprotease in Aeromonas veronii biovar sobria. J. Med. Microbiol. 2004, 53, 477–482. [Google Scholar] [CrossRef]
- Kim, J.A.; Park, J.H.; Lee, M.A.; Lee, H.J.; Park, S.J.; Kim, K.S.; Choi, S.H.; Lee, K.H. Stationary-phase induction of vvpS expression by three transcription factors: Repression by LeuO and activation by SmcR and CRP. Mol. Microbiol. 2015, 97, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Wang, D.F.; Liu, N.; Ma, Y.; Ding, T.; Mei, Y.C.; Li, J.R. Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde. Int. J. Food Microbiol. 2018, 269, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K.; Barrios, A.F.G.; Herzberg, M.; Lee, J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbial. Biotechnol. 2006, 72, 361–367. [Google Scholar] [CrossRef]
- O’May, C.; Tufenkji, N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 2011, 77, 3061–3067. [Google Scholar] [CrossRef]
- Defrancesco, A.S.; Masloboeva, N.; Syed, A.K.; DeLoughery, A.; Bradshaw, N.; Li, G.W.; Gilmore, M.S.; Walker, S.; Losick, R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, e5969–e5978. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, S.; Alghofaili, F.; Freestone, P.; Yesilkaya, H. Host stress hormone norepinephrine stimulates pneumococcal growth, biofilm formation and virulence gene expression. BMC. Microbiol. 2014, 14, 180. [Google Scholar] [CrossRef]
- Wickramasinghe, N.N.; Ravensdale, J.; Coorey, R.; Dykes, G.A.; Chandry, P.S. Transcriptional profiling of biofilms formed on chilled beef by psychrotrophic meat spoilage bacterium, Pseudomonas fragi 1793. Biofilm 2021, 3, 100045. [Google Scholar] [CrossRef] [PubMed]
- Viale, A.M.; Evans, B.A. Microevolution in the major outer membrane protein OmpA of Acinetobacter baumannii. Microb. Genom. 2020, 6, 000381. [Google Scholar] [CrossRef]
- Lynch, M.J.; Swift, S.; Kirke, D.F.; Keevil, C.W.; Dodd, C.E.R.; Williams, P. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 2002, 4, 18–28. [Google Scholar] [CrossRef]
- Kozlova, E.V.; Khajanchi, B.K.; Sha, J.A.; Chopra, A.K. Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 2011, 50, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, E.V.; Khajanchi, B.K.; Popov, V.L.; Wen, J.L.; Chopra, A.K. Impact of QseBC system in c-di-GMP-dependent quorum sensing regulatory network in a clinical isolate SSU of Aeromonas hydrophila. Microb. Pathog. 2012, 53, 115–124. [Google Scholar] [CrossRef] [PubMed]



| Primer | Sequence (5′-3′) |
|---|---|
| rpob-F | GGATCACGGTGCCTACAT |
| rpob-R | TAACGCTCGGAAGAGAAGA |
| hly F | TCCGCACTATCTTGGCATCC |
| hly R | TCTACCTCAACGTCAACCGC |
| omp-F | GCAGAACAAACCGCTCAAGG |
| omp-R | CGATCAGGGATTCGGTGGAG |
| luxR-F | GCAGACCCCGAACAAGGACACC |
| luxR-R | CACGCATGAAACCGGCAAACAG |
| luxS-F | GACTTCGGGGTGAGAAACGG |
| luxS-R | AAGATGTAGCTGGCGCAGAA |
| qseB-F | CCAGTCTTCCCGCATCACCAAC |
| qseB-R | GAGCTCCCTGTCCTCTCCCCGC |
| Concentration of Oridoninμg/mL | OD260 | |||
|---|---|---|---|---|
| 0 h | 3 h | 6 h | 9 h | |
| 0 | 0.092 ± 0.006 a | 0.104 ± 0.004 a | 0.109 ± 0.014 a | 0.126 ± 0.006 a |
| 1/4 × MIC | 0.112 ± 0.003 a | 0.391 ± 0.059 b | 0.403 ± 0.055 b | 0.419 ± 0.069 b |
| 1/2 × MIC | 0.124 ± 0.015 a | 0.532 ± 0.043 b | 0.553 ± 0.043 b | 0.561 ± 0.047 b |
| 1 × MIC | 0.141 ± 0.026 a | 1.010 ± 0.140 c | 1.014 ± 0.137 c | 1.032 ± 0.127 c |
| 2 × MIC | 0.092 ± 0.074 a | 1.822 ± 0.186 d | 1.827 ± 0.164 d | 1.832 ± 0.165 d |
| Concentration of Oridonin μg/mL | Concentration of Intracellular Liquid Protein (μg/mL) | ||
|---|---|---|---|
| 3 h | 6 h | 9 h | |
| 0 | 1075.85 ± 10.60 a | 1092.02 ± 11.32 a | 1026.64 ± 42.10 a |
| 1/4 × MIC | 936.15 ± 24.35 b | 836.71 ± 185.21 b | 1019.07 ± 123.98 a |
| 1/2 × MIC | 895.89 ± 64.169 b | 1005.31 ± 513.46 a | 880.06 ± 41.94 b |
| 1 × MIC | 671.55 ± 90.36 c | 672.58 ± 84.66 d | 745.18 ± 84.08 b |
| 2 × MIC | 669.14 ± 102.87 c | 742.43 ± 29.35 bd | 807.81 ± 21.08 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, H.; Chen, J.; Wang, Y.; Gu, Y.; Jiu, M. Antibacterial Mechanisms and Antivirulence Activities of Oridonin against Pathogenic Aeromonas hydrophila AS 1.1801. Microorganisms 2024, 12, 415. https://doi.org/10.3390/microorganisms12020415
Wang L, Li H, Chen J, Wang Y, Gu Y, Jiu M. Antibacterial Mechanisms and Antivirulence Activities of Oridonin against Pathogenic Aeromonas hydrophila AS 1.1801. Microorganisms. 2024; 12(2):415. https://doi.org/10.3390/microorganisms12020415
Chicago/Turabian StyleWang, Lunji, Huijuan Li, Jinhao Chen, Yi Wang, Yuqing Gu, and Min Jiu. 2024. "Antibacterial Mechanisms and Antivirulence Activities of Oridonin against Pathogenic Aeromonas hydrophila AS 1.1801" Microorganisms 12, no. 2: 415. https://doi.org/10.3390/microorganisms12020415





