African Swine Fever Virus I267L Is a Hemorrhage-Related Gene Based on Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and the Virus
2.2. RNA-Seq and Data Analysis
2.3. Reference Database
2.4. RNA Isolation and Quantitative Real-Time Reverse Transcription PCR(RT-qPCR)
2.5. Virus Titration
2.6. Flow Cytometry
2.7. TNF-α Stimulation
2.8. Biosafety Statement and Facility
2.9. Data Availability
3. Results
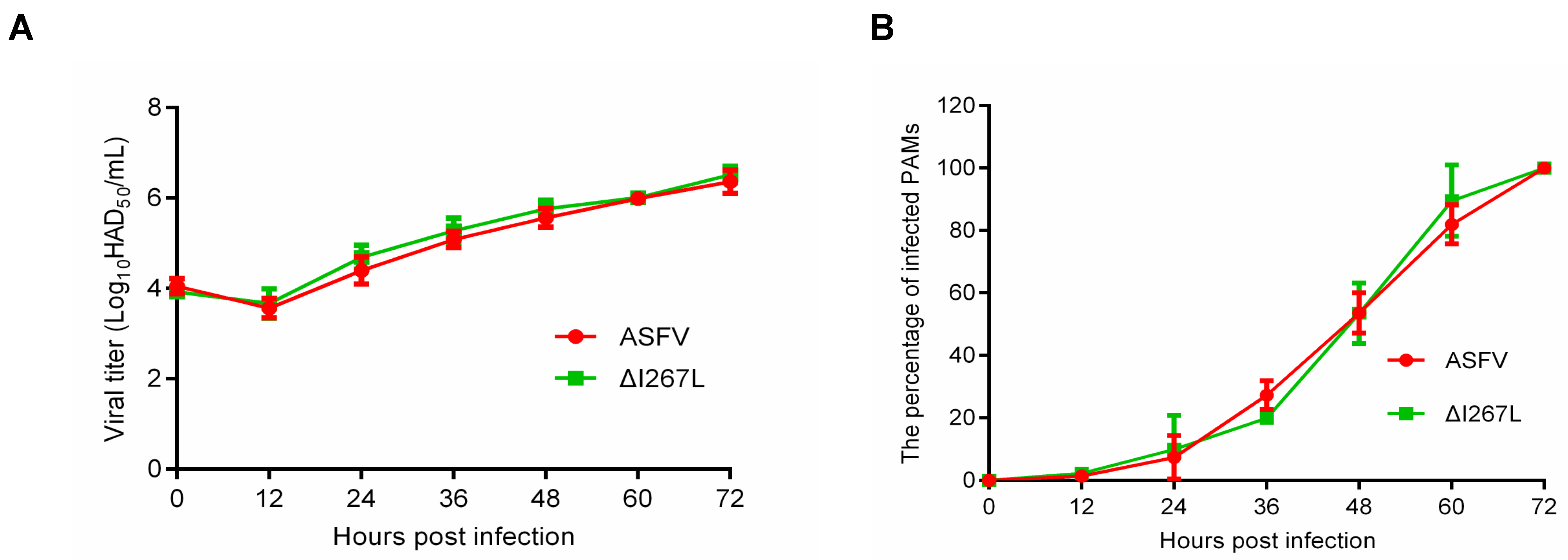
3.1. Growth Kinetic of the ΔI267L Strain
3.2. RNA-Seq and Sequencing Data Quality Analysis
3.3. Differentially Expressed Genes
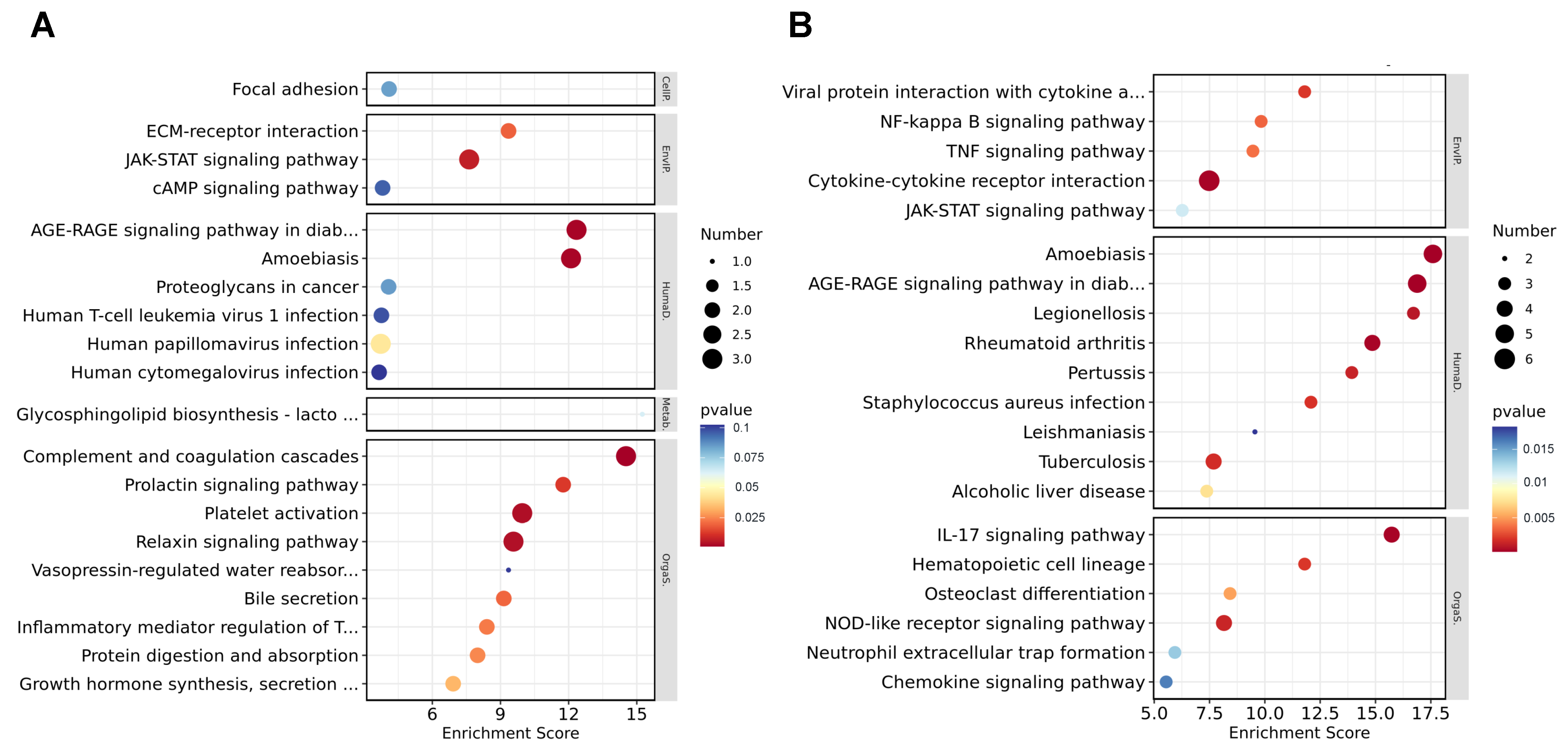
3.4. Gene Ontology Analysis of DEGs
3.5. Kyoto Encyclopedia of Genes and Genomes Analysis of DEGs
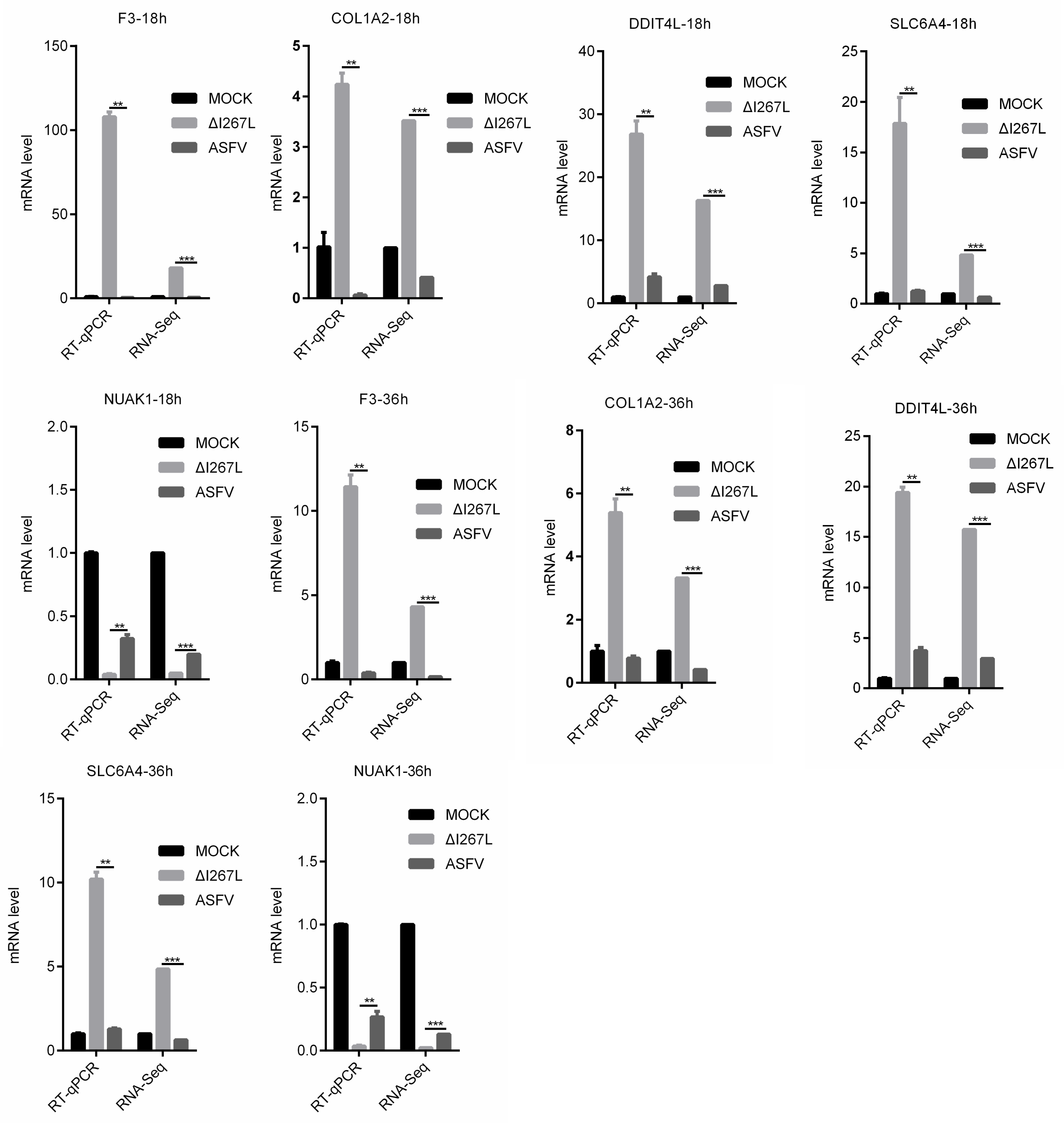
3.6. Validation of RNA-Seq Results
3.7. I267L Can Inhibit TNF-α-Induced F3 Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.L.; Bastos, A.D.; Etter, E.M.C.; Beltran-Alcrudo, D. Epidemiology of African swine fever in Africa today: Sylvatic cycle versus socio-economic imperatives. Transbound. Emerg. Dis. 2019, 66, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Cardiel, I.; Ballester, M.; Solanes, D.; Nofrarías, M.; López-Soria, S.; Argilaguet, J.M.; Lacasta, A.; Accensi, F.; Rodríguez, F.; Segalés, J. Standardization of pathological investigations in the framework of experimental ASFV infections. Virus Res. 2013, 173, 180–190. [Google Scholar] [CrossRef]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Pikalo, J.; Zani, L.; Huhr, J.; Beer, M.; Blome, S. Pathogenesis of African swine fever in domestic pigs and European wild boar—Lessons learned from recent animal trials. Virus Res. 2019, 271, 197614. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.W.; Vu, T.T.H.; Le, V.P.; Yeom, M.; Song, D.; Jeong, D.G.; Park, S.K. Advanced Strategies for Developing Vaccines and Diagnostic Tools for African Swine Fever. Viruses 2023, 15, 2169. [Google Scholar] [CrossRef]
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African Swine Fever Epidemiology and Control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef] [PubMed]
- Costard, S.; Mur, L.; Lubroth, J.; Sanchez-Vizcaino, J.M.; Pfeiffer, D.U. Epidemiology of African swine fever virus. Virus Res. 2013, 173, 191–197. [Google Scholar] [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef]
- Yutin, N.; Wolf, Y.I.; Raoult, D.; Koonin, E.V. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009, 6, 223. [Google Scholar] [CrossRef]
- Fernandez, A.; Perez, J.; Carrasco, L.; Bautista, M.J.; Sanchez-Vizcaino, J.M.; Sierra, M.A. Distribution of ASFV antigens in pig tissues experimentally infected with two different Spanish virus isolates. Zentralbl. Vet. B 1992, 39, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Perez, J.; Martin de las Mulas, J.; Carrasco, L.; Dominguez, J.; Sierra, M.A. Localization of African swine fever viral antigen, swine IgM, IgG and C1q in lung and liver tissues of experimentally infected pigs. J. Comp. Pathol. 1992, 107, 81–90. [Google Scholar] [CrossRef]
- Carrasco, L.; Gomez-Villamandos, J.C.; Bautista, M.J.; Martin de las Mulas, J.; Villeda, C.J.; Wilkinson, P.J.; Sierra, M.A. In vivo replication of African swine fever virus (Malawi ‘83) in neutrophils. Vet. Res. 1996, 27, 55–62. [Google Scholar] [PubMed]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jia, L.; Li, J.; Liu, H.; Liu, D. Pan-Genomic Analysis of African Swine Fever Virus. Virol. Sin. 2020, 35, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, W.; Qiu, Z.; Li, Y.; Fan, J.; Wu, K.; Li, X.; Zhao, M.; Ding, H.; Fan, S.; et al. African Swine Fever Virus: A Review. Life 2022, 12, 1255. [Google Scholar] [CrossRef]
- Li, D.; Yang, W.; Li, L.; Li, P.; Ma, Z.; Zhang, J.; Qi, X.; Ren, J.; Ru, Y.; Niu, Q.; et al. African Swine Fever Virus MGF-505-7R Negatively Regulates cGAS-STING-Mediated Signaling Pathway. J. Immunol. 2021, 206, 1844–1857. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Yang, W.; Li, P.; Ru, Y.; Kang, W.; Li, L.; Ran, Y.; Zheng, H. African swine fever virus protein MGF-505-7R promotes virulence and pathogenesis by inhibiting JAK1- and JAK2-mediated signaling. J. Biol. Chem. 2021, 297, 101190. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Kang, L.; Huang, L.; Zhou, S.; Hu, L.; Zheng, J.; Li, C.; Zhang, X.; He, X.; et al. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PLoS Pathog. 2021, 17, e1009733. [Google Scholar] [CrossRef]
- Rodriguez, I.; Nogal, M.L.; Redrejo-Rodriguez, M.; Bustos, M.J.; Salas, M.L. The African swine fever virus virion membrane protein pE248R is required for virus infectivity and an early postentry event. J. Virol. 2009, 83, 12290–12300. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, W.; Wen, Y.; Niu, Q.; Yang, J.; Guan, G.; Yin, H.; Zheng, H.; Li, D.; Liu, Z. The E248R protein of African swine fever virus inhibits the cGAS-STING-mediated innate immunity. Sheng Wu Gong. Cheng Xue Bao 2022, 38, 1837–1846. [Google Scholar]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, H.; Billiar, T.R.; Kroemer, G.; Kang, R. Emerging mechanisms of immunocoagulation in sepsis and septic shock. Trends Immunol. 2021, 42, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Drake, T.A.; Morrissey, J.H.; Edgington, T.S. Selective cellular expression of tissue factor in human tissues. Implications for disorders of hemostasis and thrombosis. Am. J. Pathol. 1989, 134, 1087–1097. [Google Scholar]
- Conkling, P.R.; Greenberg, C.S.; Weinberg, J.B. Tumor necrosis factor induces tissue factor-like activity in human leukemia cell line U937 and peripheral blood monocytes. Blood 1988, 72, 128–133. [Google Scholar] [CrossRef]
- Opal, S.M. The nexus between systemic inflammation and disordered coagulation in sepsis. J. Endotoxin Res. 2004, 10, 125–129. [Google Scholar] [CrossRef]
- Vallée, I.; Tait, S.W.G.; Powell, P.P. African swine fever virus infection of porcine aortic endothelial cells leads to inhibition of inflammatory responses, activation of the thrombotic state, and apoptosis. J. Virol. 2001, 75, 10372–10382. [Google Scholar] [CrossRef]
- Karalyan, Z.A.; Sargsyan, M.A.; Arzumanyan, H.H.; Kotsinyan, A.A.; Hakobyan, L.H.; Karalova, E.M.; Voskanyan, H.E. Pathomorphology of the brain in the acute form of African swine fever. Ann. Parasitol. 2017, 63, 347–352. [Google Scholar]
- Versteeg, H.H. Tissue factor as an evolutionary conserved cytokine receptor: Implications for inflammation and signal transduction. Semin. Hematol. 2004, 41, 168–172. [Google Scholar] [CrossRef]
- Cunningham, M.A.; Romas, P.; Hutchinson, P.; Holdsworth, S.R.; Tipping, P.G. Tissue factor and factor VIIa receptor/ligand interactions induce proinflammatory effects in macrophages. Blood 1999, 94, 3413–3420. [Google Scholar] [CrossRef] [PubMed]
- Camerer, E.; Gjernes, E.; Wiiger, M.; Pringle, S.; Prydz, H. Binding of Factor VIIa to tissue factor on keratinocytes induces gene expression. J. Biol. Chem. 2000, 275, 6580–6585. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Guo, Y.; Wang, X.; Wang, Z.; Sun, L.; Dai, H.; Peng, G. Innate immune escape and adaptive immune evasion of African swine fever virus: A review. Virology 2023, 587, 109878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ke, J.; Zhang, J.; Yue, H.; Chen, T.; Li, Q.; Zhou, X.; Qi, Y.; Zhu, R.; Wang, S.; et al. I267L Is Neither the Virulence- Nor the Replication-Related Gene of African Swine Fever Virus and Its Deletant Is an Ideal Fluorescent-Tagged Virulence Strain. Viruses 2021, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Li, D.; Xiong, M.G.; Liu, H.N.; Feng, T.; Shi, Z.W.; Li, Y.H.; Wu, H.N.; Wang, S.Y.; Zheng, H.X.; et al. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 2022, 18, e1010270. [Google Scholar] [CrossRef] [PubMed]
- Lindmark, E.; Siegbahn, A. Tissue factor regulation and cytokine expression in monocyte-endothelial cell co-cultures: Effects of a statin, an ACE-inhibitor and a low-molecular-weight heparin. Thromb. Res. 2002, 108, 77–84. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Santaren, J.F.; Vinuela, E. Production and titration of African swine fever virus in porcine alveolar macrophages. J. Virol. Methods 1982, 3, 303–310. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Klipper-Aurbach, Y.; Wasserman, M.; Braunspiegel-Weintrob, N.; Borstein, D.; Peleg, S.; Assa, S.; Karp, M.; Benjamini, Y.; Hochberg, Y.; Laron, Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 1995, 45, 486–490. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Salguero, F.J.; Gil, S.; Revilla, Y.; Gallardo, C.; Arias, M.; Martins, C. Cytokine mRNA expression and pathological findings in pigs inoculated with African swine fever virus (E-70) deleted on A238L. Vet. Immunol. Immunopathol. 2008, 124, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Reed LJ, M.H. A Simple Method of Estimating Fifty per Cent Endpoints 12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Neilan, J.G.; Lu, Z.; Kutish, G.F.; Zsak, L.; Burrage, T.G.; Borca, M.V.; Carrillo, C.; Rock, D.L. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitro and viral virulence. Virology 1997, 230, 252–264. [Google Scholar] [CrossRef]
- Kleinegris, M.C.; Ten Cate-Hoek, A.J.; Ten Cate, H. Coagulation and the vessel wall in thrombosis and atherosclerosis. Pol. Arch. Med. Wewn. 2012, 122, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Kay, A.M.; Simpson, C.L.; Stewart, J.A., Jr. The Role of AGE/RAGE Signaling in Diabetes-Mediated Vascular Calcification. J. Diabetes Res. 2016, 2016, 6809703. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.L.; Wang, Y.J.; Lu, Z.H.; Tian, L.; Xia, Z.Q.; Wang, K.L.; Chen, T.; Wang, R.; Feng, Z.Y.; Shi, G.P.; et al. Wumei Wan attenuates angiogenesis and inflammation by modulating RAGE signaling pathway in IBD: Network pharmacology analysis and experimental evidence. Phytomedicine 2023, 111, 154658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Dai, J.; Zhang, K.; Wang, T.; Li, L.F.; Luo, Y.; Sun, Y.; Qiu, H.J.; Li, S. The H240R Protein of African Swine Fever Virus Inhibits Interleukin 1beta Production by Inhibiting NEMO Expression and NLRP3 Oligomerization. J. Virol. 2022, 96, e0095422. [Google Scholar] [CrossRef]
- Franzoni, G.; Pedrera, M.; Sanchez-Cordon, P.J. African Swine Fever Virus Infection and Cytokine Response In Vivo: An Update. Viruses 2023, 15, 233. [Google Scholar] [CrossRef]
- Bensaude, E.; Turner, J.L.E.; Wakeley, P.R.; Sweetman, D.A.; Pardieu, C.; Drew, T.W.; Wileman, T.; Powell, P.P. Classical swine fever virus induces proinflammatory cytokines and tissue factor expression and inhibits apoptosis and interferon synthesis during the establishment of long-term infection of porcine vascular endothelial cells. J. Gen. Virol. 2004, 85, 1029–1037. [Google Scholar] [CrossRef]
- Rogers, K.J.; Maury, W. The role of mononuclear phagocytes in Ebola virus infection. J. Leukoc. Biol. 2018, 104, 717–727. [Google Scholar] [CrossRef]
- Mackman, N.; Grover, S.P.; Antoniak, S. Tissue factor expression, extracellular vesicles, and thrombosis after infection with the respiratory viruses influenza A virus and coronavirus. J. Thromb. Haemost. 2021, 19, 2652–2658. [Google Scholar] [CrossRef] [PubMed]
- Hisada, Y.; Sachetto, A.T.A.; Mackman, N. Circulating tissue factor-positive extracellular vesicles and their association with thrombosis in different diseases. Immunol. Rev. 2022, 312, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Pennings, G.J.; Reddel, C.J.; Chen, V.M.; Gnanenthiran, S.R.; Kritharides, L. Perspective: Collagen induced platelet activation the GPVI receptor as a primary target of colchicine in cardiovascular disease. Front. Cardiovasc. Med. 2023, 9, 1104744. [Google Scholar] [CrossRef] [PubMed]
- Twardowski, T.; Fertala, A.; Orgel, J.P.R.O.; Antonio, J.D.S. Type I collagen and collagen mimetics as angiogenesis promoting superpolymers. Curr. Pharm. Des. 2007, 13, 3608–3621. [Google Scholar] [CrossRef]
- Exposito, J.Y.; Valcourt, U.; Cluzel, C.; Lethias, C. The fibrillar collagen family. Int. J. Mol. Sci. 2010, 11, 407–426. [Google Scholar] [CrossRef]
- Ding, M.; Dang, W.; Liu, H.; Xu, F.; Huang, H.; Sunkang, Y.; Li, T.; Pei, J.; Liu, X.; Zhang, Y.; et al. Combinational Deletions of MGF360-9L and MGF505-7R Attenuated Highly Virulent African Swine Fever Virus and Conferred Protection against Homologous Challenge. J. Virol. 2022, 96, e0032922. [Google Scholar] [CrossRef]




| Software | Version | Parameters | Function |
|---|---|---|---|
| fastp | 0.20.1 | length_required 50 | Raw nucleotide readings used for quality control |
| RseQC | 4.0.0 | default | RNA quality control |
| fastqc | v0.11.9 | default | Raw readings used for quality assessment |
| Hisat2 | 2.1.0 | --rna-strandness rf --fr | Genome comparison |
| samtools | 1.9 | mpileup –uRf –d 1,000,000 | Analysis of sam file and bam file |
| HTseq-count | 0.11.2 | -s reverse | Gene quantification |
| DEGSeq | 1.34.1 | q value < 0.05, |log2FoldChange| > 1 | Nonbiological repetitions used for difference analysis |
| clusterProfiler | v3.10.1 | p value < 0.05 | GO analysis and KEGG analysis |
| GenBank Number | Primers | Sequences |
|---|---|---|
| AF141959.1 | porcine GAPDH-F: | ACATGGCCTCCAAGGAGTAAGA |
| porcine GAPDH-R: | GATCGAGTTGGGGCTGTGACT | |
| FR682468.2 | p30-F: | CTCCGATGAGGGCTCTTGCT |
| p30-R: | AGACGGAATCCTCAGCATCTT | |
| AY504424.1 | porcine F3-F: | CTGGAGCCACAGGCACTAC |
| porcine F3-R: | GTCACACTCCGTGTCTGTCG | |
| AB237775.1 | COL1A2-F: | CTGGTCTTGGCGGGAACTTT |
| COL1A2-R: | AGGACCAGTCTGACCAGGTT | |
| CM000819.5 | DDIT4L-F: | GAACTCCCAGCAGCGCC |
| DDIT4L-R: | TCGTTGAGGTTGGGTTCAGG | |
| CM000823.5 | SLC6A4-F: | AGAATGAATGAGCTCGCCACC |
| SLC6A4-R: | CGAATGGACGCTGACACACA | |
| CM000816.5 | NUAK1-F: | CCAGATGTGCAGTCCCCG |
| NUAK1-R: | GCAGCATTGAGGAAGCAGC | |
| CM000823.5 | COL1A1-F: | AGCCCTGGTGAAAATGGAGC |
| COL1A1-R: | AGCCCTGGTGAAAATGGAGC |
| Sample | Total Reads (M) | Clean Reads (M) | ASFV Mapped Percent | Swine Mapped Percent | Gene Number |
|---|---|---|---|---|---|
| PAM18 hpi | 50.58 | 49.8 | 0.02 | 91.19 | 14240 |
| ASFV18 hpi | 47.85 | 47.02 | 9.61 | 78.97 | 14194 |
| ΔI267L-18 hpi | 50.17 | 49.27 | 9.92 | 78.08 | 14346 |
| PAM-36 hpi | 47.9 | 47.2 | 0.02 | 91.03 | 14177 |
| ASFV36 hpi | 49.49 | 48.36 | 25.10 | 48.05 | 13724 |
| ΔI267L-36 hpi | 48.94 | 48.09 | 27.47 | 52.90 | 13888 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Duan, X.; Ren, J.; Zhang, J.; Guan, G.; Ru, Y.; Li, D.; Zheng, H. African Swine Fever Virus I267L Is a Hemorrhage-Related Gene Based on Transcriptome Analysis. Microorganisms 2024, 12, 400. https://doi.org/10.3390/microorganisms12020400
Wen Y, Duan X, Ren J, Zhang J, Guan G, Ru Y, Li D, Zheng H. African Swine Fever Virus I267L Is a Hemorrhage-Related Gene Based on Transcriptome Analysis. Microorganisms. 2024; 12(2):400. https://doi.org/10.3390/microorganisms12020400
Chicago/Turabian StyleWen, Yuan, Xianghan Duan, Jingjing Ren, Jing Zhang, Guiquan Guan, Yi Ru, Dan Li, and Haixue Zheng. 2024. "African Swine Fever Virus I267L Is a Hemorrhage-Related Gene Based on Transcriptome Analysis" Microorganisms 12, no. 2: 400. https://doi.org/10.3390/microorganisms12020400





