Native Pig Neutrophil Products: Insights into Their Antimicrobial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Peptides
2.2. Minimal Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC)
2.3. Growth Curves
2.4. Time–Kill Curves
2.5. Acridine Orange Intracellular Accumulation
2.6. Planar Lipid Bilayer Assays
2.7. Atomic Force Microscopy (AFM)
2.8. Transmission Electron Microscopy (TEM)
3. Results
3.1. Antimicrobial Activity (MIC and MBC)
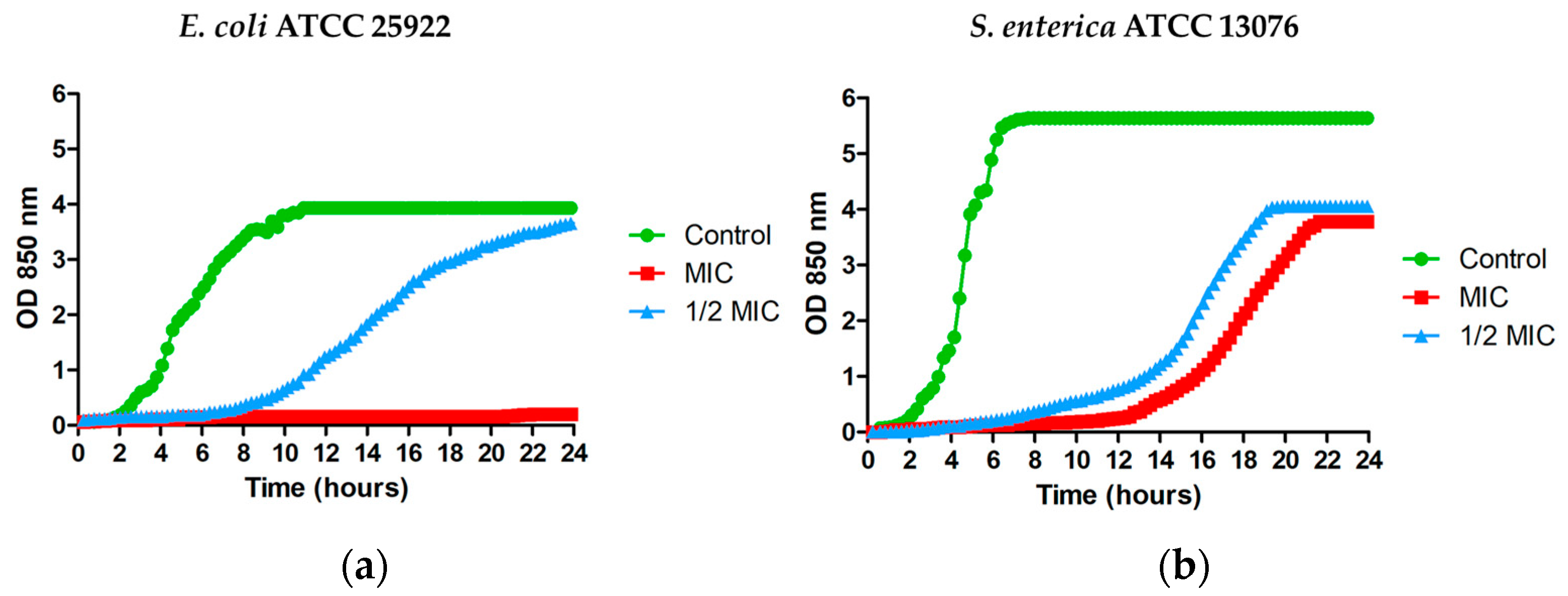
3.2. Effect of MPPN on the Microbial Growth
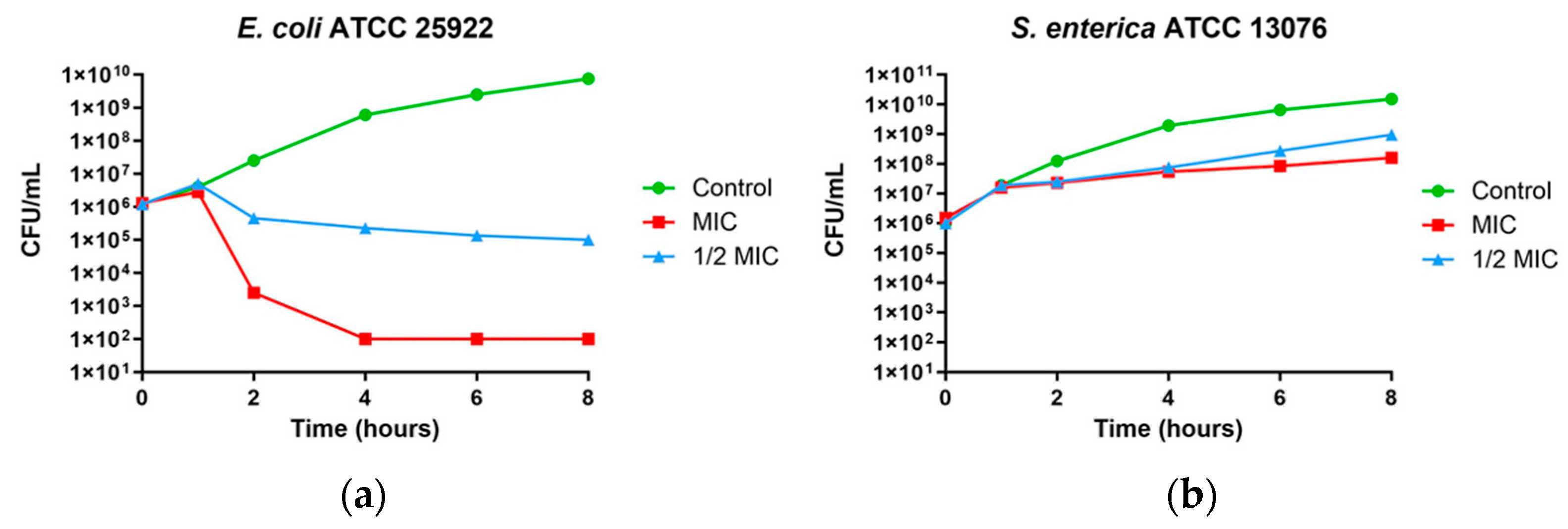
3.3. Time–Kill Curves
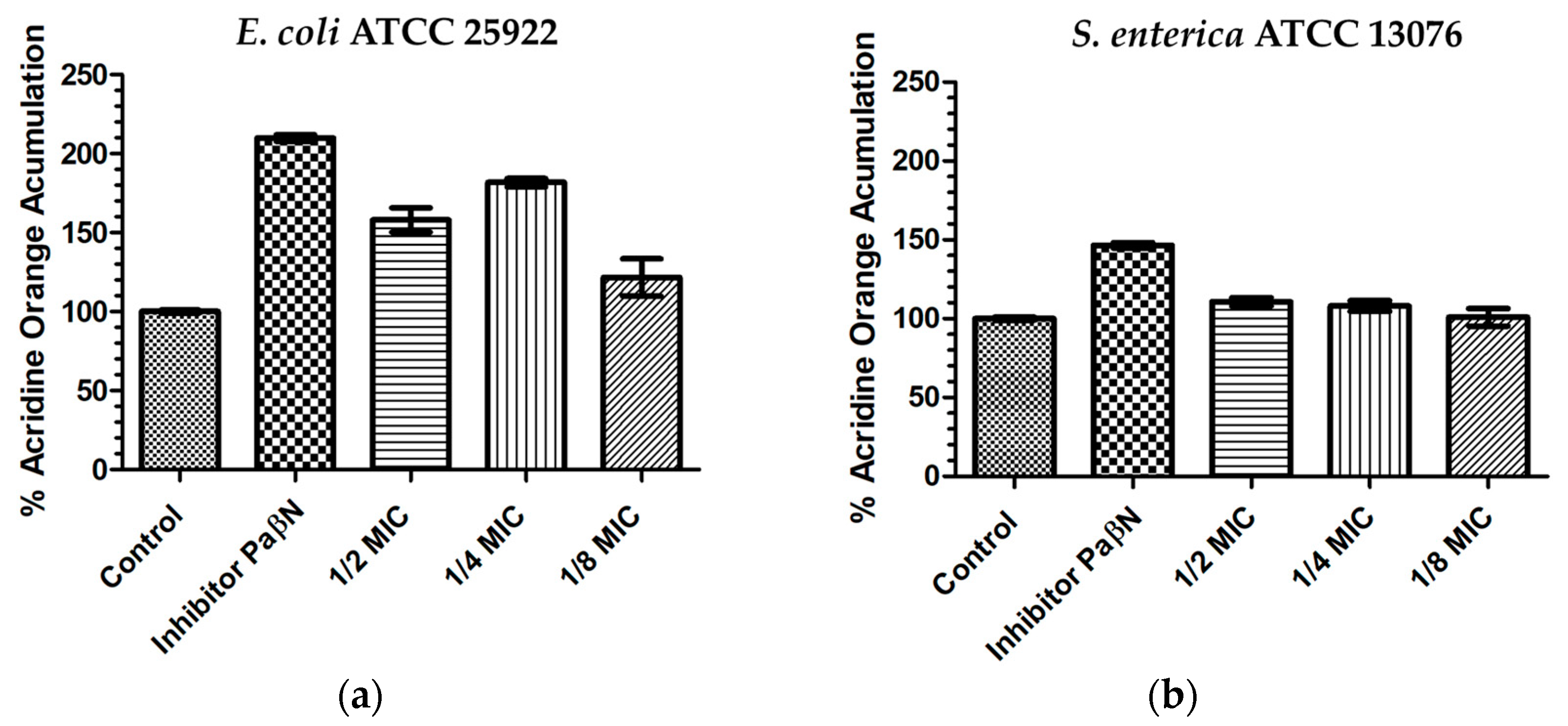
3.4. Acridine Orange Intracellular Accumulation
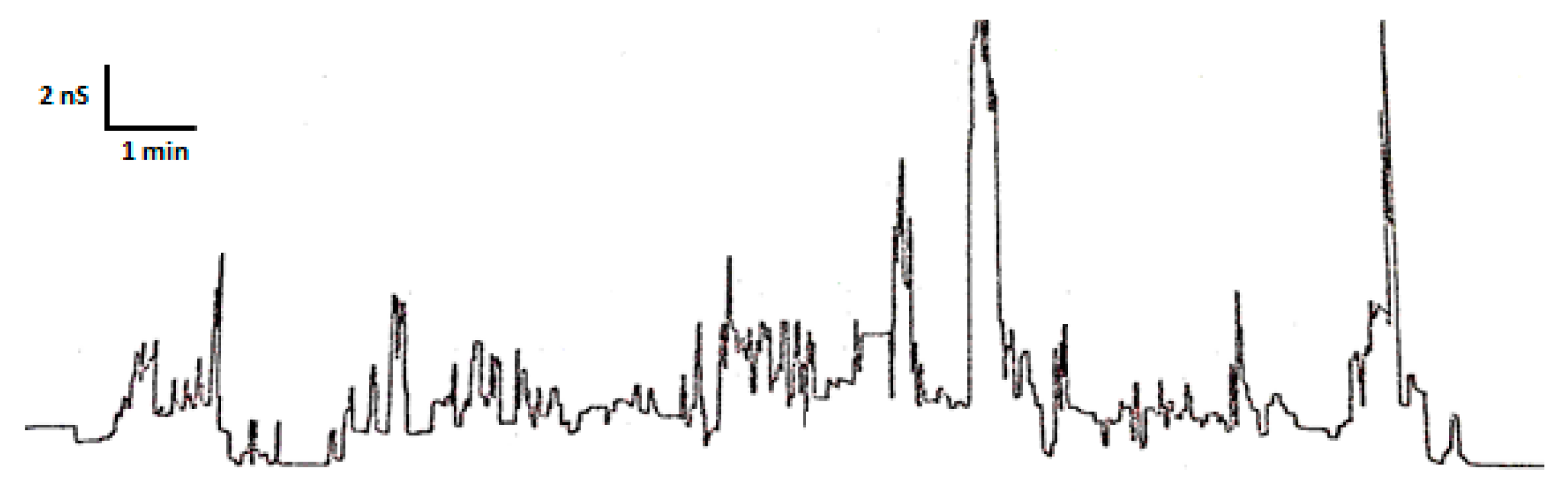
3.5. Planar Lipid Bilayer Assays
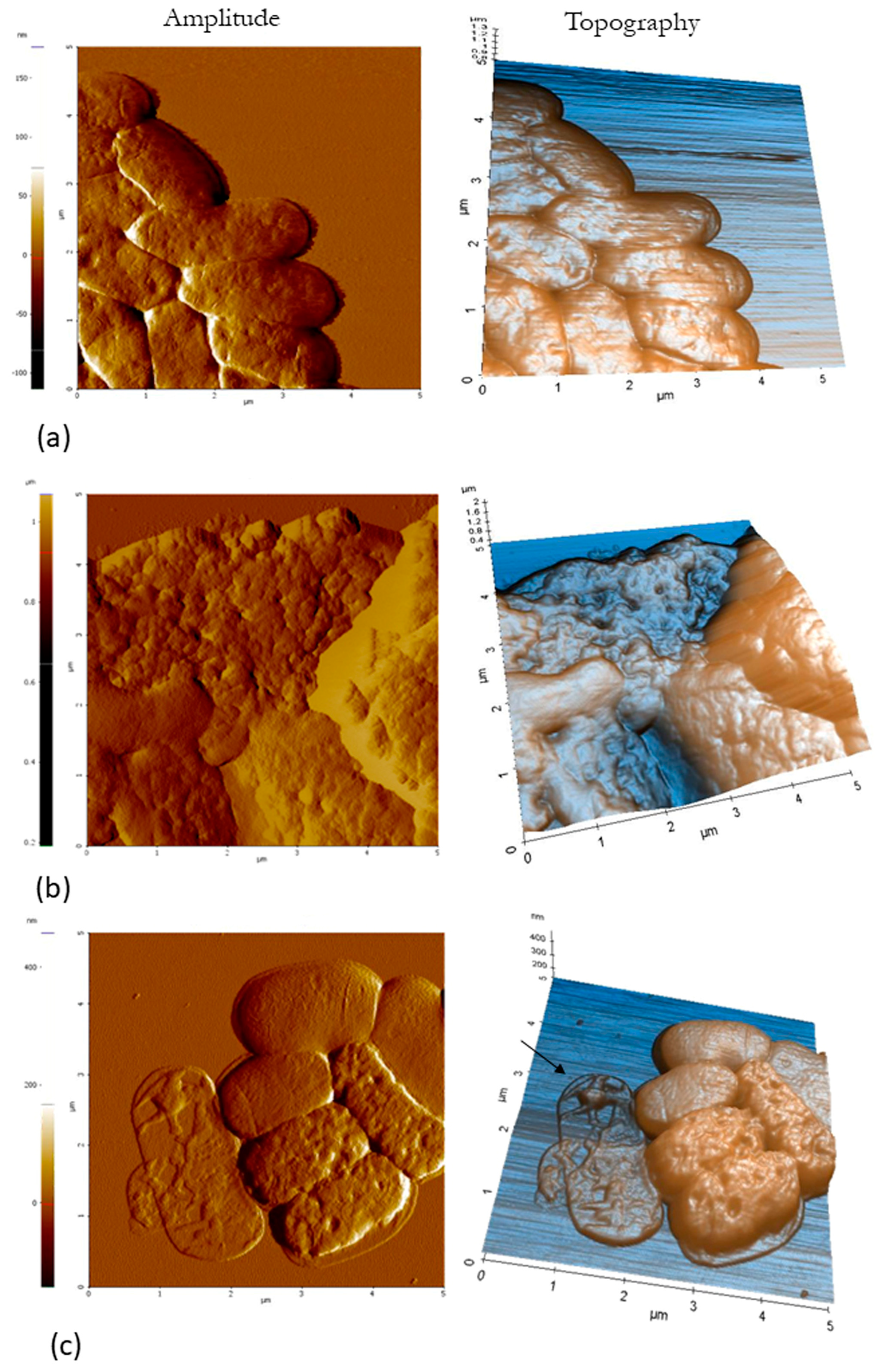
3.6. Atomic Force Microscopy (AFM)
3.7. Transmission Electron Microscopy (TEM)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Haney, E.F.; Mansour, S.C.; Hancock, R.E.W. Antimicrobial Peptides: An Introduction. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1548, pp. 3–22. [Google Scholar]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.-S.; Yang, H.; Sun, L. Antimicrobial Peptides for Combating Drug-Resistant Bacterial Infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.P.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Annunziato, G.; Costantino, G. Antimicrobial Peptides (AMPs): A Patent Review (2015–2020). Expert. Opin. Ther. Pat. 2020, 30, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro da Silva, F.; Machado, M.C.C. The Dual Role of Cathelicidins in Systemic Inflammation. Immunol. Lett. 2017, 182, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial Peptides as Therapeutic Agents: Opportunities and Challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Rollins-Smith, L.A. The Importance of Antimicrobial Peptides (AMPs) in Amphibian Skin Defense. Dev. Comp. Immunol. 2023, 142, 104657. [Google Scholar] [CrossRef]
- Bo, J.; Yang, Y.; Zheng, R.; Fang, C.; Jiang, Y.; Liu, J.; Chen, M.; Hong, F.; Bailey, C.; Segner, H.; et al. Antimicrobial Activity and Mechanisms of Multiple Antimicrobial Peptides Isolated from Rockfish Sebastiscus Marmoratus. Fish. Shellfish. Immunol. 2019, 93, 1007–1017. [Google Scholar] [CrossRef]
- Dong, N.; Chou, S.; Li, J.; Xue, C.; Li, X.; Cheng, B.; Shan, A.; Xu, L. Short Symmetric-End Antimicrobial Peptides Centered on β-Turn Amino Acids Unit Improve Selectivity and Stability. Front. Microbiol. 2018, 9, 2832. [Google Scholar] [CrossRef]
- Aarbiou, J.; Tjabringa, G.S.; Verhoosel, R.M.; Ninaber, D.K.; White, S.R.; Peltenburg, L.T.; Rabe, K.F.; Hiemstra, P.S. Mechanisms of cell death induced by the neutrophil antimicrobial peptides alpha-defensins and LL-37. Inflamm. Res. 2006, 55, 119–127. [Google Scholar] [CrossRef]
- Young-Speirs, M.; Drouin, D.; Cavalcante, P.A.; Barkema, H.W.; Cobo, E.R. Host Defense Cathelicidins in Cattle: Types, Production, Bioactive Functions and Potential Therapeutic and Diagnostic Applications. Int. J. Antimicrob. Agents 2018, 51, 813–821. [Google Scholar] [CrossRef]
- Wiesner, J.; Vilcinskas, A. Antimicrobial Peptides: The Ancient Arm of the Human Immune System. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Zheng, W.; Chen, M.; Zou, Q.; Tang, C.; Zhou, X. Genetically Engineered Pigs for Xenotransplantation: Hopes and Challenges. Front. Cell Dev. Biol. 2023, 10, 1093534. [Google Scholar] [CrossRef] [PubMed]
- Wessely-Szponder, J.; Majer-Dziedzic, B.; Smolira, A. Analysis of Antimicrobial Peptides from Porcine Neutrophils. J. Microbiol. Methods 2010, 83, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ross, C.R.; Blecha, F. Porcine antimicrobial peptides: New prospects for ancient molecules of host defense. Vet. Res. 2000, 31, 277–296. [Google Scholar] [CrossRef]
- Wessely-Szponder, J.; Bobowiec, R.; Szponder, T. The influence of porcine prophenin on neutrophils isolated from rabbit blood during implantation of calcium sulphate graft material into bone tissue. World Rabbit. Sci. 2012, 20, 163–172. [Google Scholar] [CrossRef]
- Vunnam, S.; Juvvadi, P.; Merrifield, R.B. Synthesis and Antibacterial Action of Cecropin and Proline-Arginine-Rich Peptides from Pig Intestine. J. Pept. Res. 1997, 49, 59–66. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Harwig, S.S.L.; Kokryakov, V.N.; Swiderek, K.M.; Aleshina, G.M.; Zhao, C.; Lehrer, R.I. Prophenin-1, an Exceptionally Proline-Rich Antimicrobial Peptide from Porcine Leukocytes. FEBS Lett. 1995, 362, 65–69. [Google Scholar] [CrossRef]
- Han, F.F.; Wang, Y.Z.; Feng, J.; Guo, J.; Xu, Z.R. Developmental Gene Expression of Antimicrobial Peptide Protegrin-1 and Effect of Weaning on Gene Regulation of Protegrin-1 in Piglets. J. Anim. Feed. Sci. 2007, 16, 86–95. [Google Scholar] [CrossRef]
- Anderson, R.; Wilkinson, B.; Yu, P.-L. Ovine antimicrobial peptides: New products from an age-old industry. Aust. J. Agric. Res. 2004, 55, 69–75. [Google Scholar] [CrossRef]
- Anderson, R.; Yu, P. Pilot-scale extraction and antimicrobial activity of crude extract from ovine neutrophils. Process Biochem. 2008, 43, 882–886. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Hancock, R.E.W. Function and therapeutic potential of host defence peptides. J. Pept. Sci. 2005, 11, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Głuch, K.; Bajuk, A.; Michalak, L. The time of flight mass spectrometry in investigation of high molecules. Electronics 2001, 68, 8–9. [Google Scholar]
- Gruszecka, A.; Szymańska-Chargot, M.; Smolira, A.; Michalak, L. Role of the target materials on a laser desorption/ionization mass spectra. Rapid Commun. Mass. Spectrom. 2008, 22, 925–929. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Armengol, E.; Domenech, O.; Fusté, E.; Pérez-Guillén, I.; Borrell, J.H.; Sierra, J.M.; Vinas, M. Efficacy of Combinations of Colistin with Other Antimicrobials Involves Membrane Fluidity and Efflux Machinery. Infect. Drug Resist. 2019, 12, 2031–2038. [Google Scholar] [CrossRef]
- Kordel, M.; Benz, R.; Sahll, H.-G. Mode of Action of the Staphylococcinlike Peptide Pep 5: Voltage-Dependent Depolarization of Bacterial and Artificial Membranes. J. Bacteriol. 1988, 170, 84–88. [Google Scholar] [CrossRef]
- Wu, M.; Maier, E.; Benz, R.; Hancock, R.E.W. Mechanism of Interaction of Different Classes of Cationic Antimicrobial Peptides with Planar Bilayers and with the Cytoplasmic Membrane of Escherichia coli. Biochemistry 1999, 38, 7235–7242. [Google Scholar] [CrossRef]
- Allison, D.P.; Mortensen, N.P.; Sullivan, C.J.; Doktycz, M.J. Atomic Force Microscopy of Biological Samples. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 618–634. [Google Scholar] [CrossRef]
- Sans-Serramitjana, E.; Fusté, E.; Martínez-Garriga, B.; Merlos, A.; Pastor, M.; Pedraz, J.L.; Esquisabel, A.; Bachiller, D.; Vinuesa, T.; Viñas, M. Killing Effect of Nanoencapsulated Colistin Sulfate on Pseudomonas Aeruginosa from Cystic Fibrosis Patients. J. Cyst. Fibros. 2016, 15, 611–618. [Google Scholar] [CrossRef]
- Etxeberria, M.; López-Jiménez, L.; Merlos, A.; Escuín, T.; Viñas, M. Bacterial Adhesion Efficiency on Implant Abutments: A Comparative Study. Int. Microbiol. 2013, 16, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells Are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of Antimicrobial Peptides. A Review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.M.; Shi, J.; Ceccarelli, A.; Kim, Y.-H.; Park, A.; Ganz, T. Inhibition of Neutrophil Elastase Prevents Cathelicidin Activation and Impairs Clearance of Bacteria from Wounds. Blood 2001, 97, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Scapinello, S.; Brooks, A.S.; MacInnes, J.I.; Hammermueller, J.; Clark, M.E.; Caswell, J.L. Bactericidal Activity of Porcine Neutrophil Secretions. Vet. Immunol. Immunopathol. 2011, 139, 113–118. [Google Scholar] [CrossRef]
- Ankomah, P.; Levin, B.R. Exploring the Collaboration between Antibiotics and the Immune Response in the Treatment of Acute, Self-Limiting Infections. Proc. Natl. Acad. Sci. USA 2014, 111, 8331–8338. [Google Scholar] [CrossRef]
- Strukelj, B.; Pungercar, J.; Kopitar, G.; Renko, M.; Lenarcic, B.; Berbic, S.; Turk, V. Molecular Cloning and Identification of a Novel Porcine Cathelin-Like Antibacterial Peptide Precursor. Biol. Chem. Hoppe Seyler 1995, 376, 507–510. [Google Scholar] [CrossRef]
- Zhao, C.; Ganz, T.; Lehrer, R.I. Structures of Genes for Two Cathelin-Associated Antimicrobial Peptides: Prophenin-2 and PR-39. FEBS Lett. 1995, 376, 130–134. [Google Scholar] [CrossRef]
- Gudmundsson, G.H.; Magnussont, K.P.; Chowdhary, B.P.; Johansson, M.; Andersson, L.; Boman, H.G. Structure of the Gene for Porcine Peptide Antibiotic PR-39, a Cathelin Gene Family Member: Comparative Mapping of the Locus for the Human Peptide Antibiotic FALL-39. Proc. Natl. Acad. Sci. USA 1995, 92, 7085–7089. [Google Scholar] [CrossRef]
- Fahrnerl, R.L.; Dieckmannl, T.; Harwig, S.S.; Lehrer, R.I.; Eisenbergl, D.; Feigot, J. Solution Structure of Protegrin-1, a Broad-Spectrum Antimicrobial Peptide from Porcine Leukocytes. Chem. Biol. 1996, 3, 543–550. [Google Scholar] [CrossRef]
- Kokryakov, V.N.; Harwig, S.S.L.; Panyutich, E.A.; Shevchenko, A.A.; Aleshina, G.M.; Shamova, O.V.; Korneva, H.A.; Lehrer, R.I. Protegrins: Leukocyte Antimicrobial Peptides That Combine Features of Corticostatic Defensins and Tachyplesins. FEBS Lett. 1993, 327, 231–236. [Google Scholar] [CrossRef]
- Dé, J.-F.; Sanchez, R.; Ois Hoh, F.; Strub, M.-P.; Aumelas, A.; Dumas, C. Structure of the cathelicidin motif of protegrin-3 precursor: Structural insights into the activation mechanism of an antimicrobial protein. Structure 2002, 10, 1363–1370. [Google Scholar] [CrossRef]
- Martynowycz, M.W.; Rice, A.; Andreev, K.; Nobre, T.M.; Kuzmenko, I.; Wereszczynski, J.; Gidalevitz, D. Salmonella Membrane Structural Remodeling Increases Resistance to Antimicrobial Peptide LL-37. ACS Infect. Dis. 2019, 5, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [PubMed]
- Blair, J.M.A.; Zeth, K.; Bavro, V.N.; Sancho-Vaello, E. The Role of Bacterial Transport Systems in the Removal of Host Antimicrobial Peptides in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2022, 46, fuac032. [Google Scholar] [CrossRef]
- Viñas, M.; Rabanal, F.; Benz, R.; Vinuesa, T.; Fuste, E. Perspectives in the Research on Antimicrobial Peptides. In Antimicrobial Compounds: Current Strategies and New Alternatives; Villa, T.G., Veiga-Crespo, P., Eds.; Springer Verlag: Berlin/Heidelberg, Germany, 2014; Volume 9783642404443, pp. 269–284. [Google Scholar]
- Boheim, G. Statistical Analysis of Alamethicin Channels in Black Lipid Membranes. J. Membr. Biol. 1974, 19, 277–303. [Google Scholar] [CrossRef]
- Gutsmann, T.; Riekens, B.; Bruhn, H.; Wiese, A.; Seydel, U.; Leippe, M. Interaction of Amoebapores and NK-Lysin with Symmetric Phospholipid and Asymmetric Lipopolysaccharide/Phospholipid Bilayers. Biochemistry 2003, 42, 9804–9812. [Google Scholar] [CrossRef]
- Hagge, S.O.; Wiese, A.; Seydel, U.; Gutsmann, T. Inner Field Compensation as a Tool for the Characterization of Asymmetric Membranes and Peptide-Membrane Interactions. Biophys. J. 2004, 86, 913–922. [Google Scholar] [CrossRef]
- Arcisio-Miranda, M.; dos Santos Cabrera, M.P.; Konno, K.; Rangel, M.; Procopio, J. Effects of the Cationic Antimicrobial Peptide Eumenitin from the Venom of Solitary Wasp Eumenes Rubronotatus in Planar Lipid Bilayers: Surface Charge and Pore Formation Activity. Toxicon 2008, 51, 736–745. [Google Scholar] [CrossRef]
- Domingues, M.M.; Silva, P.M.; Franquelim, H.G.; Carvalho, F.A.; Castanho, M.A.R.B.; Santos, N.C. Antimicrobial Protein RBPI21-Induced Surface Changes on Gram-Negative and Gram-Positive Bacteria. Nanomedicine 2014, 10, 543–551. [Google Scholar] [CrossRef]
- Rudilla, H.; Fusté, E.; Cajal, Y.; Rabanal, F.; Vinuesa, T.; Viñas, M. Synergistic Antipseudomonal Effects of Synthetic Peptide AMP38 and Carbapenems. Molecules 2016, 21, 1223. [Google Scholar] [CrossRef] [PubMed]

| Name ID | Peptide Name | Peptide Sequence | Type of Structure | Size, Amino Acids |
|---|---|---|---|---|
| PF-1 | Prophenin-1 | AFPPPNVPGPRFPPPNFPGPRFPPPNFPGPRFPPPNFPGPRFPPPNFPGPPFPPPIFPGPWFPPPPPFRPPPFGPPRFP | Type II poly-L-proline helix conformation | 79 |
| PF-2 | Prophenin-2 | AFPPPNVPGPRFPPPNVPGPRFPPPNFPGPRFPPPNFPGPRFPPPNFPGPPFPPPIFPGPWFPPPPPFRPPPFGPPRFP | 79 | |
| PR-39 | PR-39 | RRRPRPPYLPRPRPPPFFPPRLPPRIPPGFPPRFPPRFP-NH2 | 39 | |
| PG-1 | Protegrin-1 | RGGRLCYCRRRFCVCVGR-NH2 | Disulfide-bond-stabilized ϒ-core conformation | 18 |
| PG-2 | Protegrin-2 | RGGRLCYCRRRFCICV-NH2 | 16 | |
| PG-3 | Protegrin-3 | RGGGLCYCRRRFCVCVGR-NH2 | 18 |
| Gram-Negative | Gram-Positive | ||||||
|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | A. baumannii ATCC 17978 | P. aeruginosa ATCC 27583 | K. pneumoniae ATCC 28331 | S. enterica ATCC 13076 | E. faecalis ATCC 29212 | S. aureus ATCC 29213 | |
| MIC (μg/mL) | 68.7 | >275 | >275 | >275 | 68.7 | >275 | >275 |
| MBC (μg/mL) | 137.5 | >275 | >275 | >275 | >275 | >275 | >275 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-De La Cruz, E.; Wessely-Szponder, J.; Viñas, M.; Vinuesa, T.; Merlos, A.; Jorba, M.; Espinal, P.; Fusté, E. Native Pig Neutrophil Products: Insights into Their Antimicrobial Activity. Microorganisms 2023, 11, 2119. https://doi.org/10.3390/microorganisms11082119
Fernández-De La Cruz E, Wessely-Szponder J, Viñas M, Vinuesa T, Merlos A, Jorba M, Espinal P, Fusté E. Native Pig Neutrophil Products: Insights into Their Antimicrobial Activity. Microorganisms. 2023; 11(8):2119. https://doi.org/10.3390/microorganisms11082119
Chicago/Turabian StyleFernández-De La Cruz, Eric, Joanna Wessely-Szponder, Miguel Viñas, Teresa Vinuesa, Alexandra Merlos, Marta Jorba, Paula Espinal, and Ester Fusté. 2023. "Native Pig Neutrophil Products: Insights into Their Antimicrobial Activity" Microorganisms 11, no. 8: 2119. https://doi.org/10.3390/microorganisms11082119






