Investigating the Impact of Tillage and Crop Rotation on the Prevalence of phlD-Carrying Pseudomonas Potentially Involved in Disease Suppression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Collection and Treatment
2.3. Bacterial Strain and Culture Storage
2.4. DNA Extraction and Determining Quality–Quantity of DNA
2.5. Generation of Standard Curve and phlD Quantification in Samples
2.6. Calculating Copy Number (CN) g−1 of Soil and g−1 of Root Samples following qPCR
2.7. Statistical Analysis
3. Results
3.1. Frequency of phlD in the Rhizospheres of OSR and Wheat Crops under Different Tillage Systems
3.2. Population Density of Indigenous phlD Microbes in the Roots of OSR and Wheat Crops under Different Tillage Systems
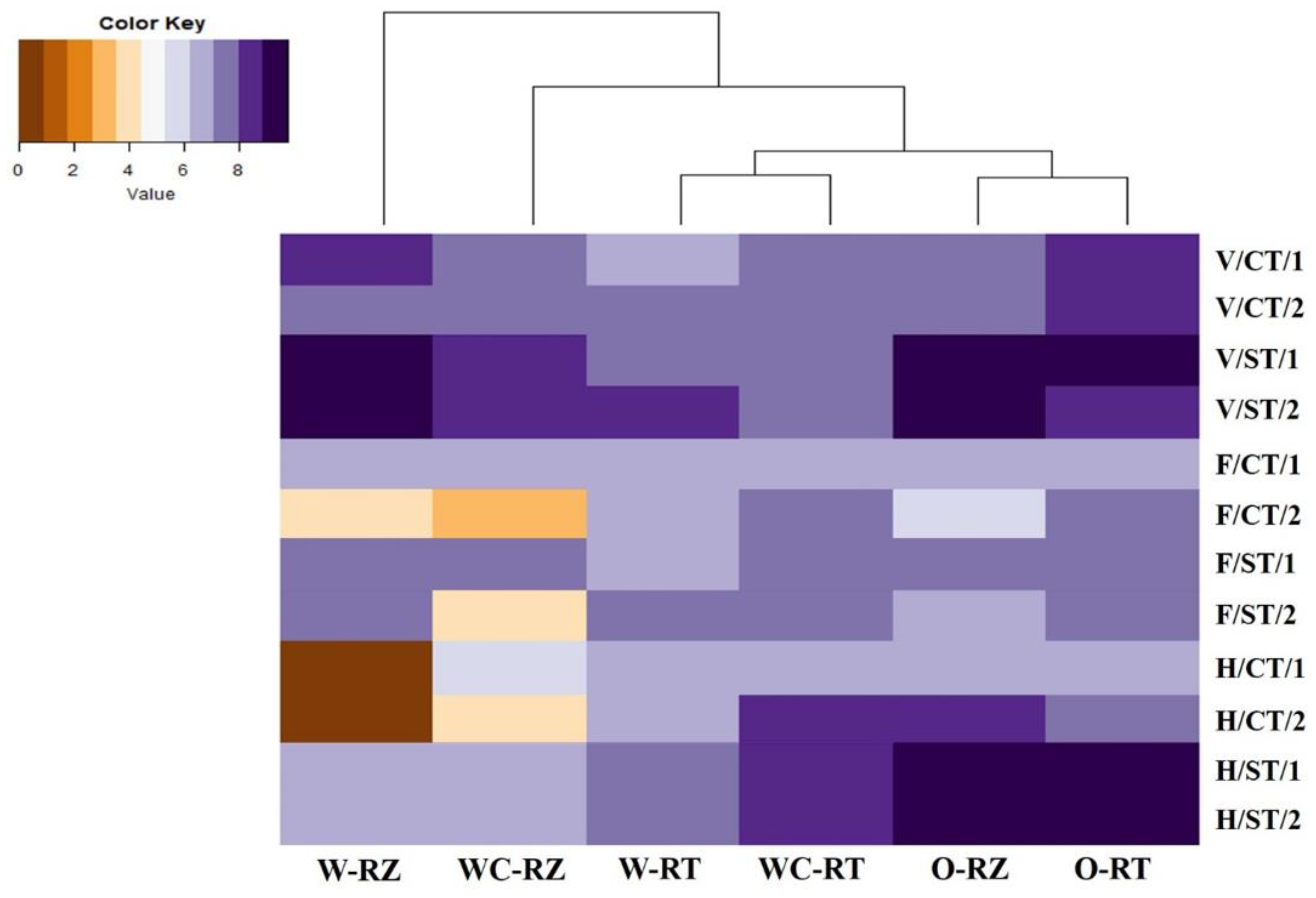
3.3. Relationships between Microhabitat Zones and Crops of Antimicrobial Gene phlD Abundance at Different Plant Growth Stages in Two Continuous Years
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lamichhane, J.R.; Corrales, D.C.; Soltani, E. Biological seed treatments promote crop establishment and yield: A global meta-analysis. Agron. Sustain. Dev. 2022, 42, 45. [Google Scholar] [CrossRef]
- Rotenberg, D.; Joshi, R.; Benitez, M.-S.; Chapin, L.G.; Camp, A.; Zumpetta, C.; Osborne, A.; Dick, W.A.; Gardener, B.B.M.; Khoa, N.; et al. Farm Management Effects on Rhizosphere Colonization by Native Populations of 2,4-Diacetylphloroglucinol-Producing Pseudomonas spp. and Their Contributions to Crop Health. Phytopathology 2007, 97, 756–766. [Google Scholar] [CrossRef]
- Picard, C.; Bosco, M. Heterozygosis drives maize hybrids to select elite 2,4-diacethylphloroglucinol-producing Pseudomonas strains among resident soil populations. FEMS Microbiol. Ecol. 2006, 58, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.B.; Lutz, M.P.; Frapolli, M.; Péchy-Tarr, M.; Rochat, L.; Keel, C.; Défago, G.; Maurhofer, M. Interplay between Wheat Cultivars, Biocontrol Pseudomonads, and Soil. Appl. Environ. Microbiol. 2010, 76, 6196–6204. [Google Scholar] [CrossRef]
- Frapolli, M.; Défago, G.; Moënne-Loccoz, Y. Denaturing gradient gel electrophoretic analysis of dominant 2,4-diacetylphloroglucinol biosynthetic phlD alleles in fluorescent Pseudomonas from soils suppressive or conducive to black root rot of tobacco. Soil Biol. Biochem. 2010, 42, 649–656. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.V.; Cappellari, I.R.; Giordano, W.; Banchio, E. Plant growth-promoting effects of native Pseudomonas strains on Mentha piperita (peppermint): An in vitro study. Plant Biol. 2015, 17, 1218–1226. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Diversity and Natural Functions of Antibiotics Produced by Beneficial and Plant Pathogenic Bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Singh, M.; Singh, D.; Gupta, A.; Pandey, K.D.; Singh, P.K.; Kumar, A. Chapter Three—Plant Growth Promoting Rhizo-bacteria: Application in Biofertilizers and Biocontrol of Phytopathogens. In PGPR Amelioration in Sustainable Agriculture; Singh, A.K., Kumar, A., Singh, P.K., Eds.; Woodhead Publishing: Cambridge, UK, 2019; pp. 41–66. ISBN 9780128158791. [Google Scholar]
- Wang, H.; Liu, R.; You, M.P.; Barbetti, M.J.; Chen, Y. Pathogen Biocontrol Using Plant Growth-Promoting Bacteria (PGPR): Role of Bacterial Diversity. Microorganisms 2021, 9, 1988. [Google Scholar] [CrossRef]
- Keerthana, U.; Prabhukarthikeyan, S.R.; Baite, M.S.; Yadav, M.K.; Kumar, R.N.; Kumar, M.A.; Raghu, S.; Aravindan, S.; Rath, P.C. CHAPTER 6—Fluorescent Pseudomonads: A multifaceted biocontrol agent for sustainable agriculture. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, H., Vaishnav, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 83–92. ISBN 9780323851633. [Google Scholar] [CrossRef]
- Müller, T.; Behrendt, U. Exploiting the biocontrol potential of plant-associated pseudomonads—A step towards pesticide-free agriculture? Biol. Control. 2021, 155, 104538. [Google Scholar] [CrossRef]
- Weller, D.M. Pseudomonas Biocontrol Agents of Soilborne Pathogens: Looking Back Over 30 Years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef]
- Iavicoli, A.; Boutet, E.; Buchala, A.; Métraux, J.-P.; Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R.; et al. Induced Systemic Resistance in Arabidopsis thaliana in Response to Root Inoculation with Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 2003, 16, 851–858. [Google Scholar] [CrossRef]
- Suresh, P.; Varathraju, G.; Shanmugaiah, V.; Almaary, K.S.; Elbadawi, Y.B.; Mubarak, A. Partial purification and characterization of 2,4-diacetylphloroglucinol producing Pseudomonas fluorescens VSMKU3054 against bacterial wilt disease of tomato. Saudi J. Biol. Sci. 2021, 28, 2155–2167. [Google Scholar] [CrossRef]
- Redondo-Nieto, M.; Barret, M.; Morrisey, J.P.; Germaine, K.; Martinez-Granero, F.; Barahona, E.; Navazo, A.; Sanchez-Contreras, M.; Moynihan, J.A.; Giddens, S.R.; et al. Genome Sequence of the Biocontrol Strain Pseudomonas fluorescens F113. J. Bacteriol. 2012, 194, 1273–1274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, B.; Wu, X.; Zhang, L.-Q. Characterization the role of GacA-dependent small RNAs and RsmA family proteins on 2,4-diacetylphloroglucinol production in Pseudomonas fluorescens 2P24. Microbiol. Res. 2020, 233, 126391. [Google Scholar] [CrossRef] [PubMed]
- Okubara, P.A.; Bonsall, R.F. Accumulation of Pseudomonas-derived 2,4-diacetylphloroglucinol on wheat seedling roots is influenced by host cultivar. Biol. Control. 2008, 46, 322–331. [Google Scholar] [CrossRef]
- Kwak, Y.-S.; Bonsall, R.F.; Okubara, P.A.; Paulitz, T.C.; Thomashow, L.S.; Weller, D.M. Factors impacting the activity of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens against take-all of wheat. Soil Biol. Biochem. 2012, 54, 48–56. [Google Scholar] [CrossRef]
- Gutiérrez-García, K.; Neira-González, A.; Pérez-Gutiérrez, R.M.; Granados-Ramírez, G.; Zarraga, R.; Wrobel, K.; Barona-Gómez, F.; Flores-Cotera, L.B. Phylogenomics of 2,4-Diacetylphloroglucinol-Producing Pseudomonas and Novel Antiglycation Endophytes from Piper auritum. J. Nat. Prod. 2017, 80, 1955–1963. [Google Scholar] [CrossRef]
- Bangera, M.G.; Thomashow, L.S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999, 181, 3155–3163. [Google Scholar] [CrossRef]
- Svercel, M.; Hamelin, J.; Duffy, B.; Moënne-Loccoz, Y.; Défago, G. Distribution of Pseudomonas populations harboring phlD or hcnAB biocontrol genes is related to depth in vineyard soils. Soil Biol. Biochem. 2010, 42, 466–472. [Google Scholar] [CrossRef]
- Almario, J.; Moënne-Loccoz, Y.; Muller, D. Monitoring of the relation between 2,4-diacetylphloroglucinol-producing Pseudomonas and Thielaviopsis basicola populations by real-time PCR in tobacco black root-rot suppressive and conducive soils. Soil Biol. Biochem. 2013, 57, 144–155. [Google Scholar] [CrossRef]
- Suresh, P.; Vellasamy, S.; Almaary, K.S.; Dawoud, T.M.; Elbadawi, Y.B. Fluorescent pseudomonads (FPs) as a potential biocontrol and plant growth promoting agent associated with tomato rhizosphere. J. King Saud Univ.-Sci. 2021, 33, 101423. [Google Scholar] [CrossRef]
- Burrows, M.; Sepúlveda, R.P.; Moya-Elizondo, B.R.; Ernesto, A. Integration between Pseudomonas protegens strains and fluquinconazole for the control of take-all in wheat. Crop Protection 2019, 121, 163–172. [Google Scholar]
- Patel, J.K.; Archana, G. Engineered production of 2,4-diacetylphloroglucinol in the diazotrophic endophytic bacterium Pseudomonas sp. WS5 and its beneficial effect in multiple plant-pathogen systems. Appl. Soil Ecol. 2018, 124, 34–44. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Cui, S.; Jagadamma, S.; Zhang, Q.P. Residue retention and minimum tillage improve physical environment of the soil in croplands: A global meta-analysis. Soil Tillage Res. 2019, 194, 104292. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Weller, D.M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 1998, 11, 144–152. [Google Scholar] [CrossRef]
- Mavrodi, O.V.; Mavrodi, D.V.; Parejko, J.A.; Thomashow, L.S.; Weller, D.M. Irrigation Differentially Impacts Populations of Indigenous Antibiotic-Producing Pseudomonas spp. in the Rhizosphere of Wheat. Appl. Environ. Microbiol. 2012, 78, 3214–3220. [Google Scholar] [CrossRef]
- Ownley, B.H.; Duffy, B.K.; Weller, D.M. Identification and Manipulation of Soil Properties to Improve the Biological Control Performance of Phenazine-Producing Pseudomonas fluorescens. Appl. Environ. Microbiol. 2003, 69, 3333–3343. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Durán, D.; Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment—The AmrZ-FleQ Hub. Microorganisms 2023, 11, 1037. [Google Scholar] [CrossRef]
- Cook, R.J. Take-all of wheat. Physiol. Mol. Plant Path. 2003, 62, 73–86. [Google Scholar] [CrossRef]
- McMillan, V.E.; Hammond-Kosack, K.E.; Gutteridge, R.J. Evidence that wheat cultivars differ in their ability to build up inoculum of the take-all fungus, Gaeumannomyces graminis var. tritici, under a first wheat crop. Plant Pathol. 2011, 60, 200–206. [Google Scholar] [CrossRef]
- Vrtilek, P.; Smutny, V.; Dryšlova, T.; Neudert, L.; Kren, J. The effect of agronomic measures on grain yield of winter wheat in drier conditions. Plant Soil Environ. 2018, 65, 63–70. [Google Scholar] [CrossRef]
- Kirkegaard, J.; Christen, O.; Krupinsky, J.; Layzell, D. Break crop benefits in temperate wheat production. Field Crops Res. 2008, 107, 185–195. [Google Scholar] [CrossRef]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease Suppressive Soils: New Insights from the Soil Microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- de Souza, J.T.; Weller, D.M.; Raaijmakers, J.M. Frequency, Diversity, and Activity of 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. in Dutch Take-all Decline Soils. Phytopathology 2003, 93, 54–63. [Google Scholar] [CrossRef]
- Landa, B.B.; Mavrodi, O.V.; Raaijmakers, J.M.; McSpadden Gardener, B.B.; Thomashow, L.S.; Weller, D.M. Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl. Environ. Microbiol. 2002, 68, 3226–3237. [Google Scholar] [CrossRef]
- Picard, C.; Di Cello, F.; Ventura, M.; Fani, R.; Guckert, A. Frequency and biodiversity of 2,4-Diacetylphloroglucinol producing bacteria isolated from the Maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 2000, 66, 948–955. [Google Scholar] [CrossRef]
- Brucker, R.M.; Baylor, C.M.; Walters, R.L.; Lauer, A.; Harris, R.N.; Minbiole, K.P.C. The Identification of 2,4-diacetylphloroglucinol as an Antifungal Metabolite Produced by Cutaneous Bacteria of the Salamander Plethodon cinereus. J. Chem. Ecol. 2008, 34, 39–43. [Google Scholar] [CrossRef]
- Tapia, M.P.C.; Burrows, R.P.M.; Sepúlveda, B.R.; Concha, M.V.; Palma, C.V.; Moya-Elizondo, E.A. Antagonistic Activity of Chilean Strains of Pseudomonas protegens against Fungi Causing Crown and Root Rot of Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 951. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Weller, D.M.; Thomashow, L.S. Frequency of Antibiotic-Producing Pseudomonas spp. in Natural Environments. Appl. Environ. Microbiol. 1997, 63, 881–887. [Google Scholar] [CrossRef]
- Hansen, M.L.; He, Z.; Wibowo, M.; Jelsbak, L. A Whole-Cell Biosensor for Detection of 2,4-Diacetylphloroglucinol (DAPG)-Producing Bacteria from Grassland Soil. Appl. Environ. Microbiol. 2021, 87, e01400-20. [Google Scholar] [CrossRef] [PubMed]
- Vian, J.F.; Peigne, J.; Chaussod, R.; Roger-Estrade, J. Effects of four tillage systems on soil structure and soil microbial biomass in organic farming. Soil Use Manag. 2009, 25, 1–10. [Google Scholar] [CrossRef]
- Dennert, F.; Imperiali, N.; Staub, C.; Schneider, J.; Laessle, T.; Zhang, T.; Wittwer, R.; A van der Heijden, M.G.; Smits, T.H.M.; Schlaeppi, K.; et al. Conservation tillage and organic farming induce minor variations in Pseudomonas abundance, their antimicrobial function and soil disease resistance. FEMS Microbiol. Ecol. 2018, 94, fiy075. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence (5′–3′) | Amplicon Length (bp) | Reference |
|---|---|---|---|
| B2BF | ACCCACCGCAGCATCGTTTATGAGC | 319 bp | [23] |
| B2BR3 | AGCAGAGCGACGAGAACTCCAGGGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathore, R.; Forristal, D.; Spink, J.; Dowling, D.; Germaine, K.J. Investigating the Impact of Tillage and Crop Rotation on the Prevalence of phlD-Carrying Pseudomonas Potentially Involved in Disease Suppression. Microorganisms 2023, 11, 2459. https://doi.org/10.3390/microorganisms11102459
Rathore R, Forristal D, Spink J, Dowling D, Germaine KJ. Investigating the Impact of Tillage and Crop Rotation on the Prevalence of phlD-Carrying Pseudomonas Potentially Involved in Disease Suppression. Microorganisms. 2023; 11(10):2459. https://doi.org/10.3390/microorganisms11102459
Chicago/Turabian StyleRathore, Ridhdhi, Dermot Forristal, John Spink, David Dowling, and Kieran J. Germaine. 2023. "Investigating the Impact of Tillage and Crop Rotation on the Prevalence of phlD-Carrying Pseudomonas Potentially Involved in Disease Suppression" Microorganisms 11, no. 10: 2459. https://doi.org/10.3390/microorganisms11102459





