Effect of Water Activity on Conidia Germination in Aspergillus flavus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strain and Growth Conditions
2.2. Microscopy
2.3. Flow Cytometry of Conidia
2.4. RNA-Seq Analysis
2.5. Real-Time Quantitative PCR
2.6. Tandem Mass Tag (TMT)-Labelling Analysis
2.7. Statistical Analysis
3. Results
3.1. Conidia Germination
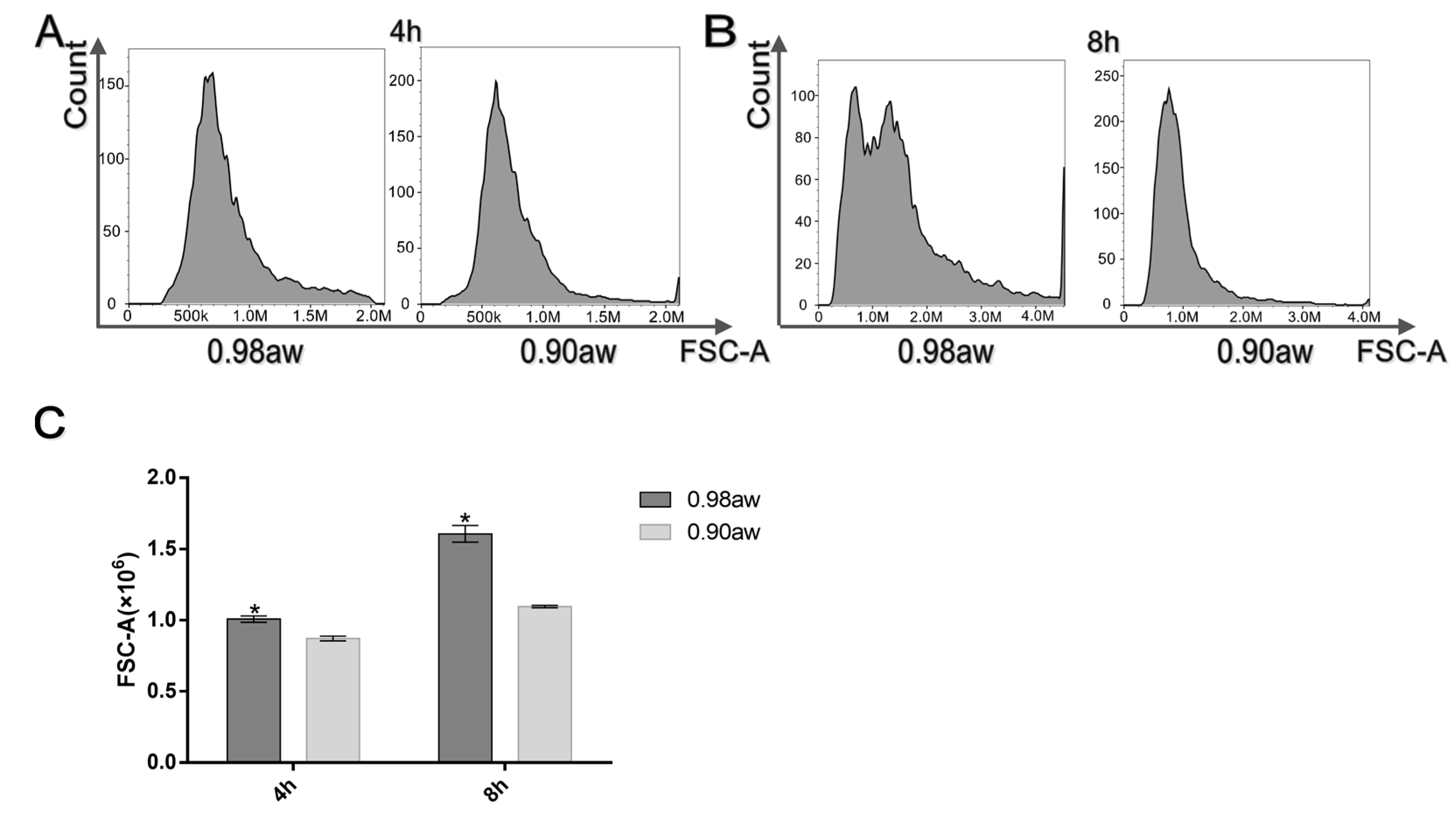
3.2. Flow Cytometry of Conidia
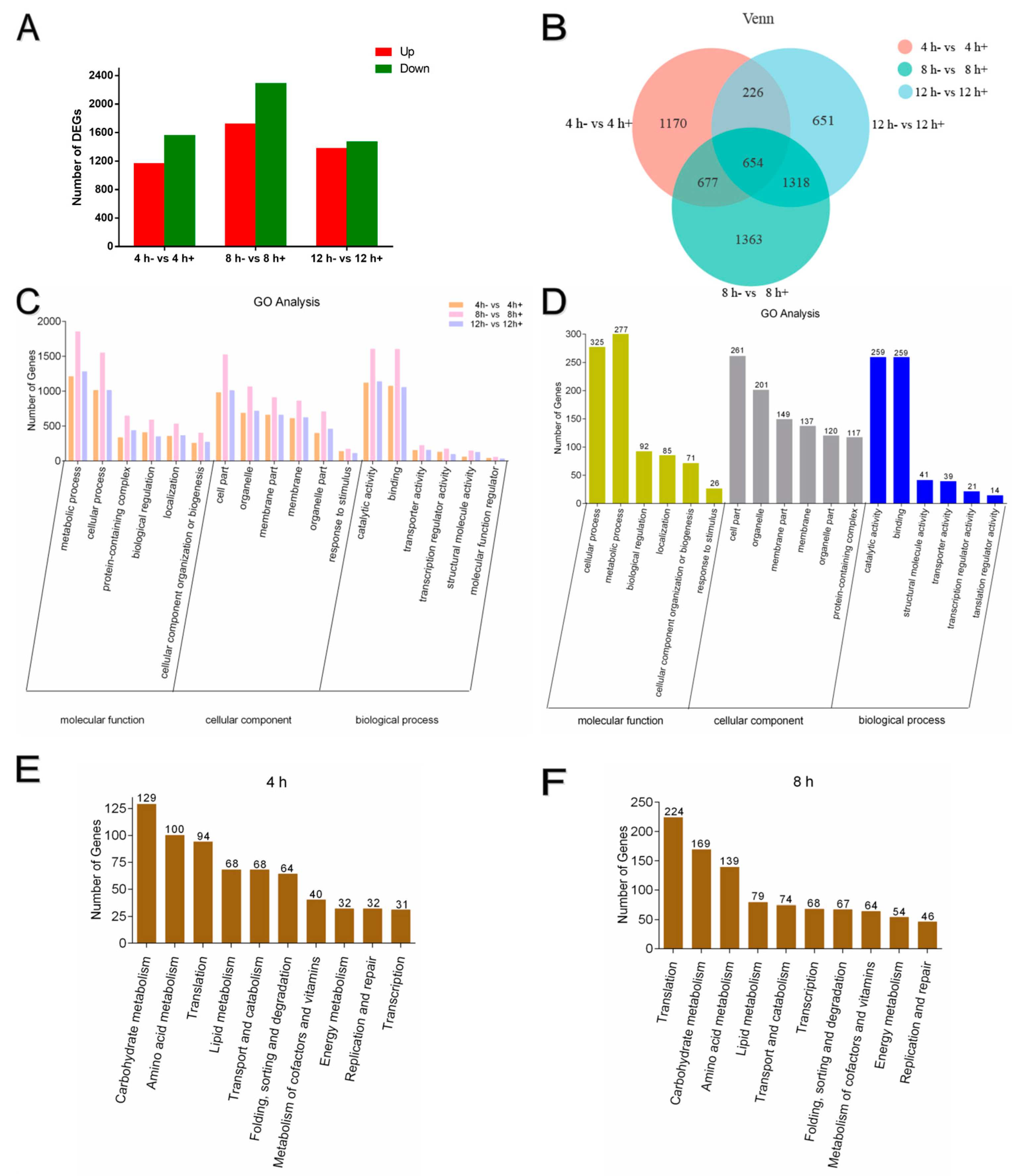
3.3. Summary of DEGs
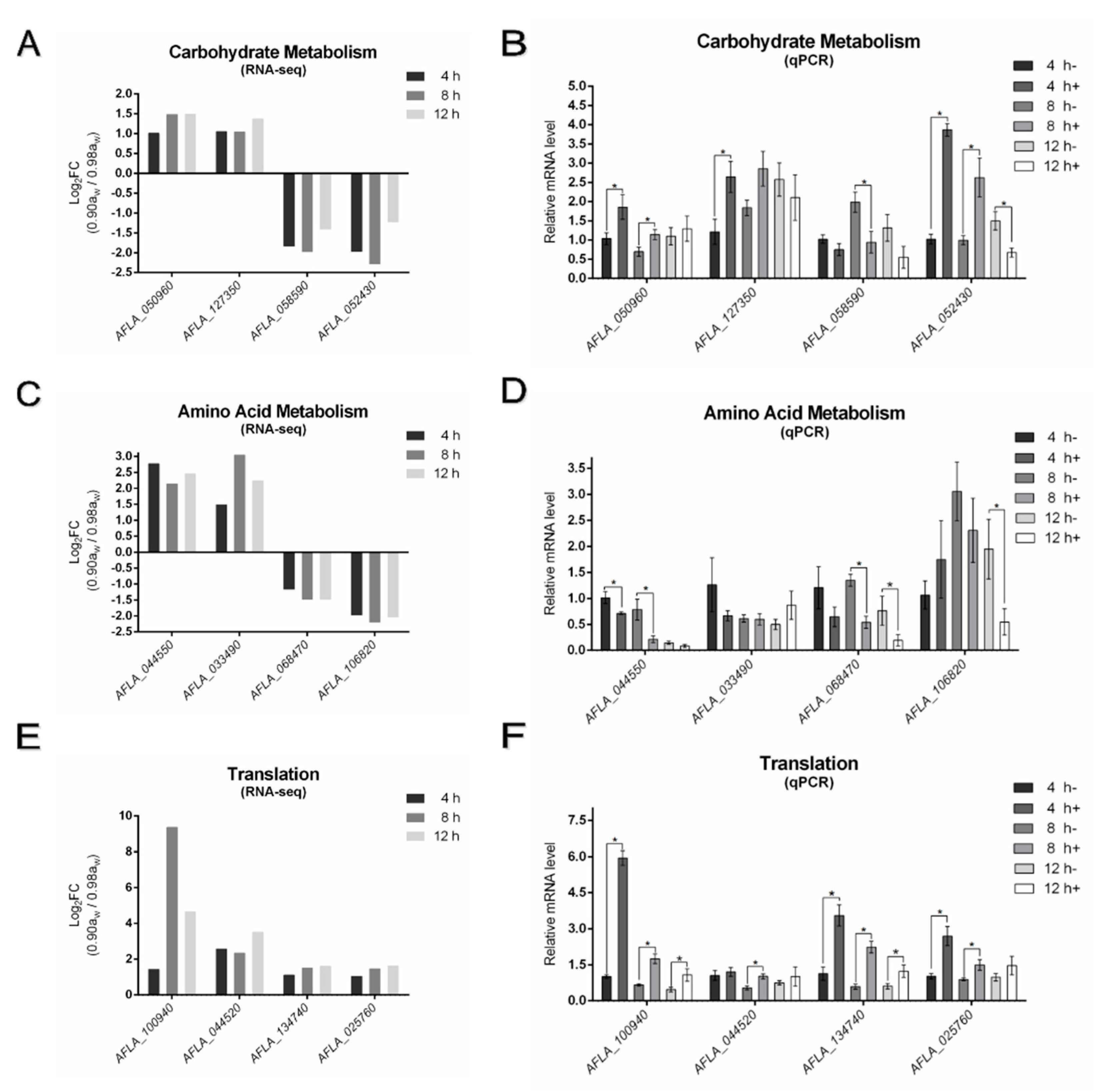
3.4. Real-Time Quantitative PCR
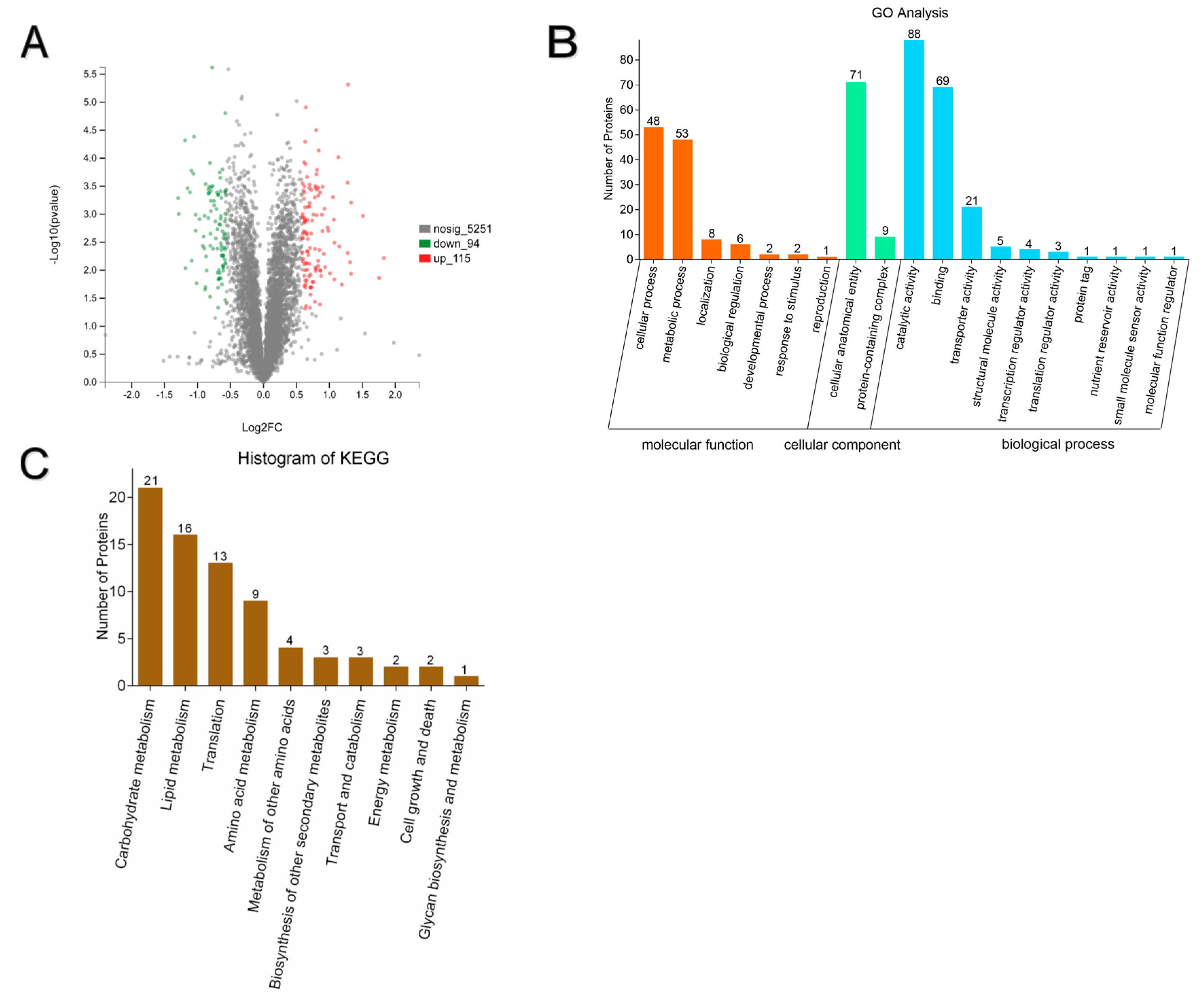
3.5. Summary of DEPs
3.6. Combined Analysis of Transcriptomics and Proteomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Microscopy
Scanning Electron Microscope
Appendix A.2. RNA-Seq Analysis
Appendix A.2.1. RNA Extraction, cDNA Library Preparation, and SEQUENCING
Appendix A.2.2. Read Mapping, Differential Expression Analysis and Functional Enrichment
Appendix A.2.3. Alternative Splice Events Identification
Appendix A.3. TMT Labeling Analysis
Appendix A.3.1. Total Protein Extraction, Protein Digestion and TMT Labeling
Appendix A.3.2. LC-MS/MS Analysis
Appendix A.3.3. Protein Identification
Appendix A.3.4. Statistical Analysis
References
- Yu, J.; Cleveland, T.E.; Nierman, W.C.; Bennett, J.W. Aspergillus flavus genomics: Gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 2005, 22, 194–202. [Google Scholar] [CrossRef] [PubMed]
- KLICH, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lv, Y.; Li, X.; Lin, Y.; Deng, H.; Pan, L. Profiling of secondary metabolite gene clusters regulated by LaeA in Aspergillus niger FGSC A1279 based on genome sequencing and transcriptome analysis. Res. Microbiol. 2018, 169, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Gizachew, D.; Hsu, Y.C.; Szonyi, B.; Ting, W.E. Effect of water activity, temperature, and incubation period on fungal growth and ochratoxin A production on Nyjer seeds. Mycotoxin Res. 2019, 35, 1–8. [Google Scholar] [CrossRef]
- Belli, N.; Marin, S.; Sanchis, V.; Ramos, A.J. Influence of water activity and temperature on growth of isolates of Aspergillus section Nigri obtained from grapes. Int. J. Food Microbiol. 2004, 96, 19–27. [Google Scholar] [CrossRef]
- Cavia, M.M.; Muiño, M.A.F.; Huidobro, J.; Sancho, M. Correlation between Moisture and Water Activity of Honeys Harvested in Different Years. J. Food Sci. 2004, 69, C368–C370. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.; Schmidt-Heydt, M.; Parra, R.; Geisen, R.; Magan, N. A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. J. R. Soc. Interface 2012, 9, 757–767. [Google Scholar] [CrossRef]
- Alam, S.; Shah, H.U.; Magan, N. Effect of calcium propionate and water activity on growth and aflatoxins production by Aspergillus flavus. J. Food Sci. 2010, 75, M61–M64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, Z.; Zhong, H.; Wang, S.; Yang, W.; Liu, Y.; Wang, S. RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 2014, 6, 3187–3207. [Google Scholar] [CrossRef]
- Mohamed, S.; Mo, L.; Flint, S.; Palmer, J.; Fletcher, G.C. Effect of water activity and temperature on the germination and growth of Aspergillus tamarii isolated from “Maldive fish”. Int. J. Food Microbiol. 2012, 160, 119–123. [Google Scholar] [CrossRef]
- Santos, J.L.P.; Chaves, R.D.; Sant’Ana, A.S. Modeling the impact of water activity, pH, and calcium propionate on the germination of single spores of Penicillium paneum. LWT Food Sci. Technol. 2020, 133, 110012. [Google Scholar] [CrossRef]
- Gougouli, M.; Koutsoumanis, K.P. Modeling germination of fungal spores at constant and fluctuating temperature conditions. Int. J. Food Microbiol. 2012, 152, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Segers, F.J.; van Laarhoven, K.A.; Huinink, H.P.; Adan, O.C.; Wosten, H.A.; Dijksterhuis, J. The Indoor Fungus Cladosporium halotolerans Survives Humidity Dynamics Markedly Better than Aspergillus niger and Penicillium rubens despite Less Growth at Lowered Steady-State Water Activity. Appl. Environ. Microbiol. 2016, 82, 5089–5098. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Jin, J.; Wang, P.; Dai, X.; Liu, Y.; Zheng, M.; Xing, F. Interaction of water activity and temperature on the growth, gene expression and aflatoxin production by Aspergillus flavus on paddy and polished rice. Food Chem. 2019, 293, 472–478. [Google Scholar] [CrossRef]
- Peromingo, B.; Rodriguez, A.; Bernaldez, V.; Delgado, J.; Rodriguez, M. Effect of temperature and water activity on growth and aflatoxin production by Aspergillus flavus and Aspergillus parasiticus on cured meat model systems. Meat Sci. 2016, 122, 76–83. [Google Scholar] [CrossRef]
- van Leeuwen, M.R.; Krijgsheld, P.; Bleichrodt, R.; Menke, H.; Stam, H.; Stark, J.; Wosten, H.A.; Dijksterhuis, J. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud. Mycol. 2013, 74, 59–70. [Google Scholar] [CrossRef]
- Yang, G.; Cao, X.; Ma, G.; Qin, L.; Wu, Y.; Lin, J.; Ye, P.; Yuan, J.; Wang, S. MAPK pathway-related tyrosine phosphatases regulate development, secondary metabolism and pathogenicity in fungus Aspergillus flavus. Environ. Microbiol. 2020, 22, 5232–5247. [Google Scholar] [CrossRef]
- Zhang, F.; Zhong, H.; Han, X.; Guo, Z.; Yang, W.; Liu, Y.; Yang, K.; Zhuang, Z.; Wang, S. Proteomic profile of Aspergillus flavus in response to water activity. Fungal Biol 2015, 119, 114–124. [Google Scholar] [CrossRef]
- Tiwari, S.; Thakur, R.; Goel, G.; Shankar, J. Nano-LC-Q-TOF Analysis of Proteome Revealed Germination of Aspergillus flavus Conidia is Accompanied by MAPK Signaling and Cell Wall Modulation. Mycopathologia 2016, 181, 769–786. [Google Scholar] [CrossRef]
- Schmidt, H.; Vlaic, S.; Kruger, T.; Schmidt, F.; Balkenhol, J.; Dandekar, T.; Guthke, R.; Kniemeyer, O.; Heinekamp, T.; Brakhage, A.A. Proteomics of Aspergillus fumigatus Conidia-containing Phagolysosomes Identifies Processes Governing Immune Evasion. Mol. Cell. Proteom. 2018, 17, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- de Assis, L.J.; Ries, L.N.; Savoldi, M.; Dinamarco, T.M.; Goldman, G.H.; Brown, N.A. Multiple Phosphatases Regulate Carbon Source-Dependent Germination and Primary Metabolism in Aspergillus nidulans. G3 (Bethesda) 2015, 5, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, T.; Coolen, J.; Zoll, J.; Verweij, P.E.; Melchers, W. Gene co-expression analysis identifies gene clusters associated with isotropic and polarized growth in Aspergillus fumigatus conidia. Fungal Genet. Biol. 2018, 116, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Herman, P.K.; Rine, J. Yeast spore germination: A requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 1997, 16, 6171–6181. [Google Scholar] [CrossRef]
- Schmidt-Heydt, M.; Abdel-Hadi, A.; Magan, N.; Geisen, R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009, 135, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG database. In Novartis Foundation Symposia; Wiley: Hoboken, NJ, USA, 2002; Volume 247, pp. 91–103; 119–128; 244–252. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Luo, G.; Zhao, L.; Xu, X.; Qin, Y.; Huang, L.; Su, Y.; Zheng, W.; Yan, Q. Integrated dual RNA-seq and dual iTRAQ of infected tissue reveals the functions of a diguanylate cyclase gene of Pseudomonas plecoglossicida in host-pathogen interactions with Epinephelus coioides. Fish Shellfish Immunol. 2019, 95, 481–490. [Google Scholar] [CrossRef]
- Osherov, N.; May, G.S. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 2001, 199, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, M.; Miskei, M.; Karanyi, Z.; Lenkey, B.; Pocsi, I.; Emri, T. Transcriptome changes initiated by carbon starvation in Aspergillus nidulans. Microbiology 2013, 159, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Hayer, K.; Stratford, M.; Archer, D.B. Structural features of sugars that trigger or support conidial germination in the filamentous fungus Aspergillus niger. Appl. Environ. Microbiol. 2013, 79, 6924–6931. [Google Scholar] [CrossRef]
- Trow, J.A. Processing of GPI-Anchored Cell Wall Proteins in Saccharomyces cerevisiae and a Role for DCW1. Ph.D. Thesis, The Johns Hopkins University, Baltimore, MD, USA, 2013. [Google Scholar]
- Alam, S.; Shah, H.U.; Magan, N. Water availability affects extracellular hydrolytic enzyme production by Aspergillus flavus and Aspergillus parasiticus. World Mycotoxin J. 2009, 2, 313–322. [Google Scholar] [CrossRef]
- Dowzer, C.E.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 1991, 11, 5701–5709. [Google Scholar] [CrossRef] [PubMed]
- Chulkin, A.M.; Vavilova, E.A.; Benevolenskii, S.V. The mutational analysis of carbon catabolite repression in filamentous fungus Penicillium canescens. Mol. Biol. 2011, 45, 871–878. [Google Scholar] [CrossRef]
- Fasoyin, O.E.; Wang, B.; Qiu, M.; Han, X.; Chung, K.R.; Wang, S. Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in Aspergillus flavus. Fungal Genet. Biol. 2018, 115, 41–51. [Google Scholar] [CrossRef]
- Fountain, J.C.; Bajaj, P.; Pandey, M.; Nayak, S.N.; Yang, L.; Kumar, V.; Jayale, A.S.; Chitikineni, A.; Zhuang, W.; Scully, B.T.; et al. Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 2016, 6, 38747. [Google Scholar] [CrossRef]
- Chen, H.; Mulder, L.; Wijma, H.J.; Wabeke, R.; Heinemann, M. A photo-switchable yeast isocitrate dehydrogenase to control metabolic flux through the citric acid cycle. bioRxiv 2021. [Google Scholar] [CrossRef]
- Palmer, J.M.; Perrin, R.M.; Dagenais, T.R.T.; Keller, N.P. H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 2052–2060. [Google Scholar] [CrossRef]
- Strahl, B.D.; Briggs, S.D.; Brame, C.J.; Caldwell, J.A.; Koh, S.S.; Ma, H.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; Stallcup, M.R.; et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001, 11, 996–1000. [Google Scholar] [CrossRef]
- Satterlee, T.; Cary, J.W.; Calvo, A.M. RmtA, a Putative Arginine Methyltransferase, Regulates Secondary Metabolism and Development in Aspergillus flavus. PLoS ONE 2016, 11, e155575. [Google Scholar] [CrossRef]
- Xu, W.; Gavia, D.J.; Tang, Y. Biosynthesis of fungal indole alkaloids. Nat. Prod. Rep. 2014, 31, 1474–1487. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.R.; Yu, J.; Bland, J.M.; Scheffler, B.E.; Kim, H.S.; Nierman, W.C.; Cleveland, T.E. Influence of tryptophan on aflatoxin biosynthesis and regulation in Aspergillus flavus. In Proceedings of the American Society for Microbiology Annual Meeting, Orlando, FL, USA, 21–26 May 2006; Abstract #O-042. p. 429. [Google Scholar]
- Grandgenett, D.P.; Stahly, D.P. Control of diaminopimelate decarboxylase by L-lysine during growth and sporulation of Bacilluscereus. J. Bacteriol. 1971, 106, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Yang, M.; Yue, Y.; Ge, F.; Li, Y.; Guo, X.; Zhang, J.; Zhang, F.; Nie, X.; Wang, S. Lysine Succinylation Contributes to Aflatoxin Production and Pathogenicity in Aspergillus flavus. Mol. Cell. Proteom. 2018, 17, 457–471. [Google Scholar] [CrossRef]
- Bhabhra, R.; Richie, D.L.; Kim, H.S.; Nierman, W.C.; Fortwendel, J.; Aris, J.P.; Rhodes, J.C.; Askew, D.S. Impaired ribosome biogenesis disrupts the integration between morphogenesis and nuclear duplication during the germination of Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 575–583. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Hawkridge, A.M.; Muddiman, D.C.; Payne, G.A. Temperature-dependent regulation of proteins in Aspergillus flavus: Whole organism stable isotope labeling by amino acids. J. Proteome Res. 2008, 7, 2973–2979. [Google Scholar] [CrossRef]
- Zhang, Y.; Wolf, G.W.; Bhat, K.; Jin, A.; Allio, T.; Burkhart, W.A.; Xiong, Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003, 23, 8902–8912. [Google Scholar] [CrossRef]
- Strudwick, S.; Borden, K.L. The emerging roles of translation factor eIF4E in the nucleus. Differentiation 2002, 70, 10–22. [Google Scholar] [CrossRef]
- Shahbazian, D.; Parsyan, A.; Petroulakis, E.; Hershey, J.; Sonenberg, N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 2010, 9, 4106–4109. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, S.; Li, C.; Wu, K.; Qi, D.; Wang, S. Effect of Water Activity on Conidia Germination in Aspergillus flavus. Microorganisms 2022, 10, 1744. https://doi.org/10.3390/microorganisms10091744
Jia S, Li C, Wu K, Qi D, Wang S. Effect of Water Activity on Conidia Germination in Aspergillus flavus. Microorganisms. 2022; 10(9):1744. https://doi.org/10.3390/microorganisms10091744
Chicago/Turabian StyleJia, Sifan, Chong Li, Kuntan Wu, Desheng Qi, and Shuai Wang. 2022. "Effect of Water Activity on Conidia Germination in Aspergillus flavus" Microorganisms 10, no. 9: 1744. https://doi.org/10.3390/microorganisms10091744




