Characterization of the Asian Citrus Psyllid-‘Candidatus Liberibacter Asiaticus’ Pathosystem in Saudi Arabia Reveals Two Predominant CLas Lineages and One Asian Citrus Psyllid Vector Haplotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Asian Citrus Psyllid Samples and DNA Extraction
2.2. Amplification and Sequencing of ACP mtCOI, Nuclear atox1, and Wolbachia Genes
2.3. ‘Candidatus Liberibacter Asiaticus’ Detection in Citrus Leaf Samples
2.4. Phylogeographic Analyses of the Asian Citrus Psyllid Mitochondrial and Nuclear Gene and Wolbachia Wsp Gene Sequences
2.5. Agilent SureSelect XT HS Target Enrichment Library Construction and Sequencing
2.6. Prophages Phylogenetic Analyses
3. Results
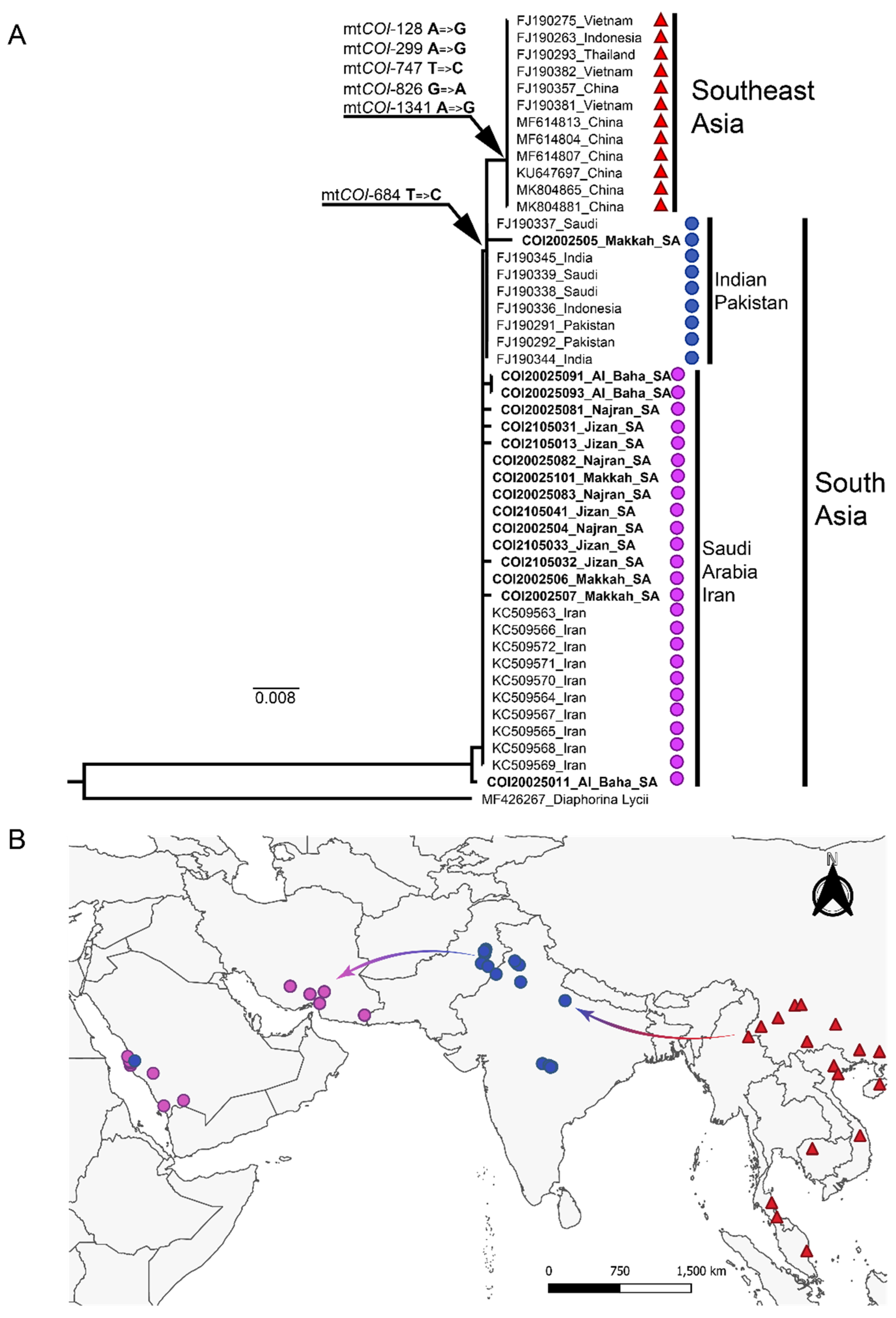
3.1. Phylogeographic Analysis of the Asian Citrus Psyllid mtCOI Sequence
3.2. Phylogenetic Analysis of the Asian Citrus Psyllid atox1 Nuclear Sequence
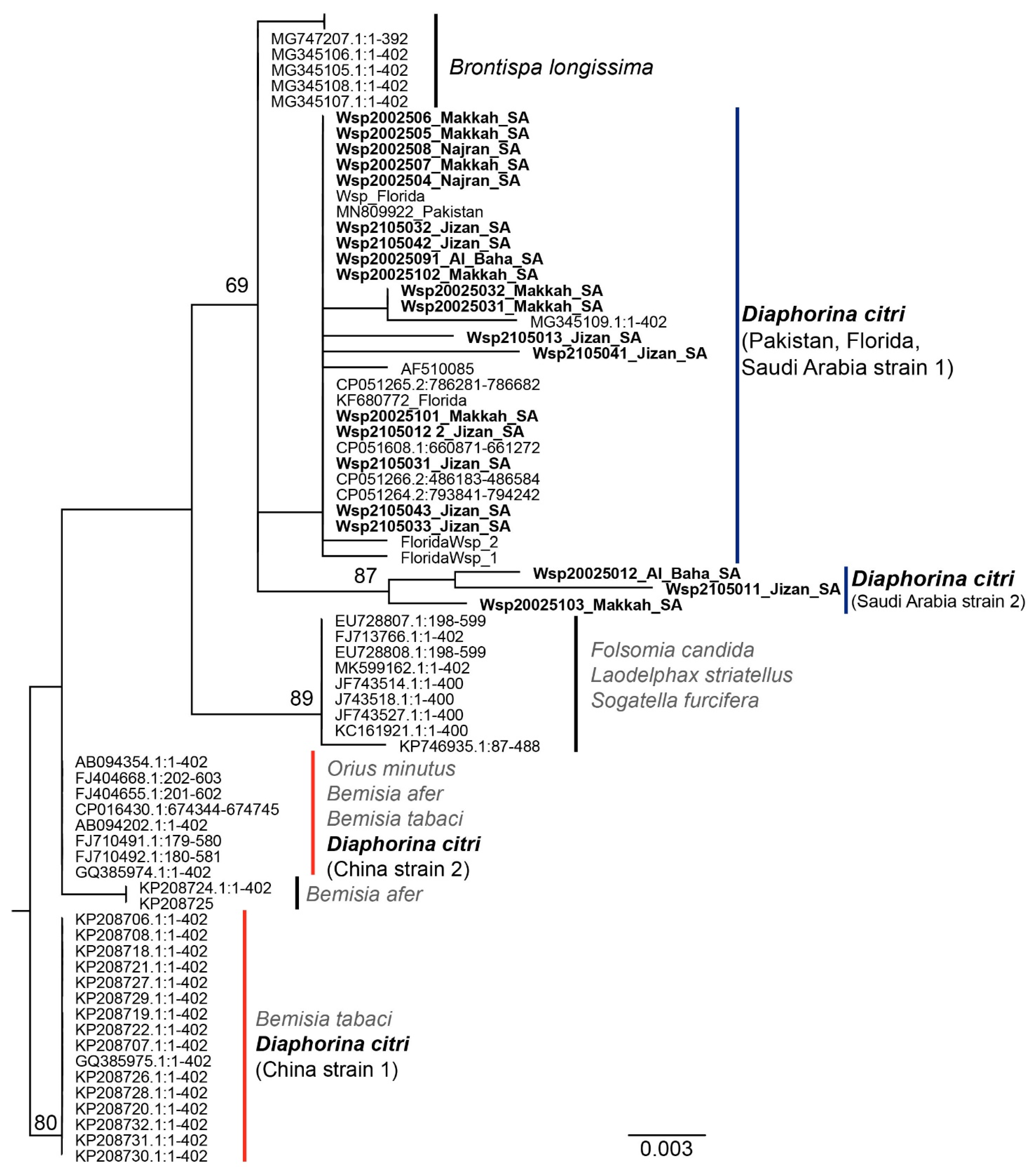
3.3. Phylogenetic Analysis of Wolbachia spp. Wsp Sequences
3.4. Analysis of ‘Candidatus Liberibacter Asiaticus’ 16S rDNA Sequence from Citrus Leaf Samples
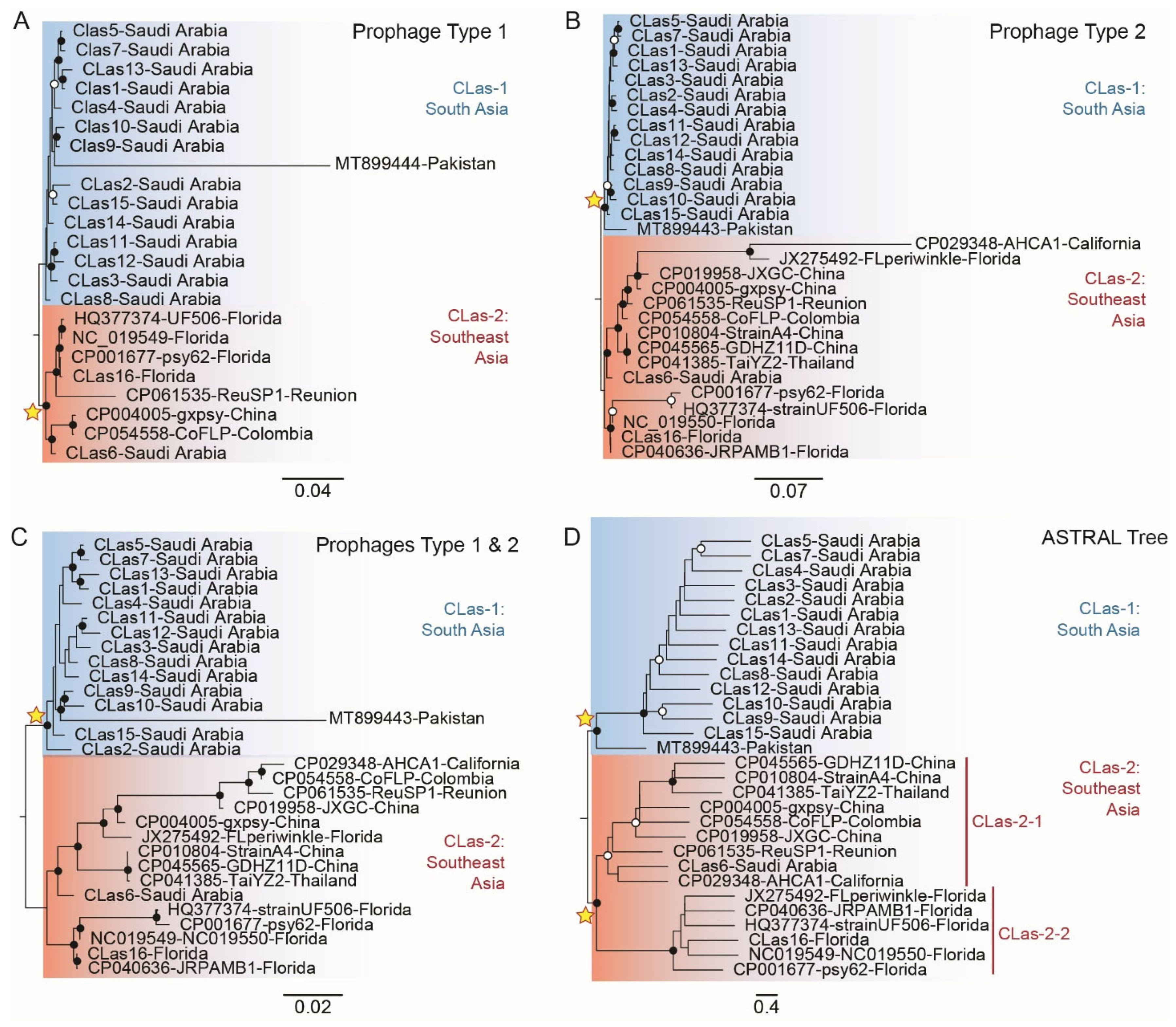
3.5. Phylogenetic Analysis of Full-Length CLas-Prophage Genome Sequences
3.6. Phylogeny of CLas Reconstructed for the SC1/type 1 Prophage Full-Length Sequences
3.7. Phylogeny of CLas Reconstructed for the SC2/type 2 Prophage Full-Length Sequences
3.8. Phylogeny of CLas Reconstructed from SC1/Type 1 and SC2/Type 2 Concatenated Genome Sequences
3.9. Reconciled CLas Species Tree Given by SC1/Type 1 and SC2/Type 2 Orthologous Genes
4. Discussion
4.1. Invasion History of Asian Citrus Psyllid in Saudi Arabia
4.2. Invasion History of ‘Candidatus Liberibacter Asiaticus’ in Saudi Arabia
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hall, D.G.; Richardson, M.L.; Ammar, E.-D.; Halbert, S.E. Asian citrus psyllid, Diaphorina citri, vector of citrus huanglongbing disease. Entomol. Exp. Appl. 2013, 146, 207–223. [Google Scholar] [CrossRef]
- Brown, J.K.; Cicero, J.M.; Fisher, T.W. Psyllid-transmitted Candidatus Liberibacter species infecting citrus and solanaceous hosts. In Vector-Mediated Transmission of Plant Pathogens; Brown, J.K., Joseph, M., Tonja, C., Fisher, W., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2016; pp. 399–422. [Google Scholar]
- Lin, K.H. Yellow shoot of citrus. Symptomatology. Investigations in the cause of huanglongbing. Natural transmission and spread. General conclusions. Acta Phytopathol. Sin. 1956, 2, 1–42. [Google Scholar]
- Batool, A.; Iftikhar, Y.; Mughal, S.M.; Khan, M.M.; Jaskani, M.J.; Abbas, M.; Khan, I.A. Citrus Greening Disease—A major cause of citrus decline in the world: A Review. Hortic. Sci. 2007, 34, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Kuwayama, S. Die psylliden Japans. Trans. Sopporo Nat. Hist. Soc. 1908, 2, 149–189. [Google Scholar]
- Reinking, O.A. Diseases of economic plants in southern China. Philipp. Agric. 1919, 8, 109–134. [Google Scholar]
- Wooler, A.; Padgham, D.; Arafat, A. Saudi Arabia-Diaphorina citri on citrus. FAO Plant Prot. Bull. 1975, 22, 93–94. [Google Scholar]
- Bové, J.M.; Garnier, M. Citrus greening and psylla vectors of the disease in the Arabian Peninsula. In Ninth International Organization of Citrus Virologists Conference Proceedings; International Organization of Citrus Virologists: Riverside, CA, USA, 1984; pp. 109–114. [Google Scholar]
- Halbert, S.E.; Manjunath, K.L. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: A literature review and assessment of risk in Florida. Fla. Entomol. 2004, 87, 330–353. [Google Scholar] [CrossRef]
- Chu, C.; Hoffmann, M.; Braswell, W.E.; Pelz-Stelinski, K.S. Genetic variation and potential coinfection of Wolbachia among widespread Asian citrus psyllid (Diaphorina citri Kuwayama) populations. Insect Sci. 2019, 26, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, C.A.; Saunders, N.C. Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu. Rev. Ecol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Frohlich, D.R.; Torres-Jerez, I.; Bedford, I.D.; Markham, P.G.; Brown, J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999, 8, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Kumagai, L.; Cen, Y.; Chen, J.; Wallis, C.M.; Polek, M.; Jiang, H.; Zheng, Z.; Liang, G.; Deng, X. Analyses of Mitogenome Sequences Revealed that Asian Citrus Psyllids (Diaphorina citri) from California Were Related to Those from Florida. Sci. Rep. 2017, 7, 10154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadjistylli, M.; Roderick, G.K.; Brown, J.K. Global population structure of a worldwide pest and virus vector: Genetic diversity and population history of the Bemisia tabaci sibling species group. PLoS ONE 2016, 11, e0165105. [Google Scholar] [CrossRef] [Green Version]
- Lavrinienko, A.; Kesäniemi, J.; Watts, P.C.; Serga, S.; Pascual, M.; Mestres, F.; Kozeretska, I. First record of the invasive pest Drosophila suzukii in Ukraine indicates multiple sources of invasion. J. Pest Sci. 2017, 90, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Paredes-Montero, J.R.; Haq, Q.M.I.; Mohamed, A.A.; Brown, J.K. Phylogeographic and SNPs analyses of Bemisia tabaci B mitotype populations reveal only two of eight haplotypes are invasive. Biology 2021, 10, 1048. [Google Scholar] [CrossRef] [PubMed]
- León, J.H.; Sétamou, M.; Gastaminza, G.A.; Buenahora, J.; Cáceres, S.; Yamamoto, P.T.; Bouvet, J.P.; Logarzo, G.A. Two separate introductions of Asian citrus psyllid populations found in the American continents. Ann. Entomol. Soc. Am. 2011, 104, 1392–1398. [Google Scholar] [CrossRef]
- Boykin, L.; De Barro, P.; Hall, D.; Hunter, W.; McKenzie, C.; Powell, C.; Shatters, R. Overview of worldwide diversity of Diaphorina citri Kuwayama mitochondrial cytochrome oxidase 1 haplotypes: Two Old World lineages and a New World invasion. Bull. Entomol. Res. 2012, 102, 573–582. [Google Scholar] [CrossRef] [Green Version]
- Guidolin, A.S.; Cônsoli, F.L. Molecular Characterization of Wolbachia Strains Associated with the Invasive Asian Citrus Psyllid Diaphorina citri in Brazil. Microb. Ecol. 2013, 65, 475–486. [Google Scholar] [CrossRef]
- Lashkari, M.; Manzari, S.; Sahragard, A.; Malagnini, V.; Boykin, L.M.; Hosseini, R. Global genetic variation in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae) and the endosymbiont Wolbachia: Links between Iran and the USA detected. Pest Manag. Sci. 2014, 70, 1033–1040. [Google Scholar] [CrossRef]
- Luo, Y.; Agnarsson, I. Global mtDNA genetic structure and hypothesized invasion history of a major pest of citrus, Diaphorina citri (Hemiptera: Liviidae). Ecol. Evol. 2018, 8, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, A.; Braswell, W.E.; Ruiz-Arce, R.; Racelis, A. Genetic variation and population structure of Diaphorina citri using cytochrome oxidase I sequencing. PLoS ONE 2018, 13, e0198399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qasim, M.; Baohua, W.; Zou, H.; Lin, Y.; Dash, C.K.; Bamisile, B.S.; Hussain, M.; Zhiwen, Z.; Wang, L. Phylogenetic relationship and genetic diversity of citrus psyllid populations from China and Pakistan and their associated Candidatus bacterium. Mol. Phylogenetics Evol. 2018, 126, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Fatima, R.; Mushtaq, S.; Salman, H.M.; Talha, M.; Razaq, S.; Haider, M.S. Molecular characterization of Asian citrus psyllid (Diaphorina citri) using mitochondrial cytochrome oxidase 1 (mtCO1) gene from Punjab Pakistan. World J. Biol. Biotechnol. 2018, 3, 203–207. [Google Scholar] [CrossRef]
- Hua, H.; Günther, V.; Georgiev, O.; Schaffner, W. Distorted copper homeostasis with decreased sensitivity to cisplatin upon chaperone Atox1 deletion in Drosophila. BioMetals 2011, 24, 445–453. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Zhang, W.; Guo, L.R. Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc. R. Soc. Lond. B Biol. Sci. 1995, 261, 55–63. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Paredes-Montero, J.R.; Zia-Ur-Rehman, M.; Hameed, U.; Haider, M.S.; Herrmann, H.W.; Brown, J.K. Genetic variability, community structure, and horizontal transfer of endosymbionts among three Asia II-Bemisia tabaci mitotypes in Pakistan. Ecol. Evol. 2020, 10, 2928–2943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanaei, E.; Charlat, S.; Engelstädter, J. Wolbachia host shifts: Routes, mechanisms, constraints and evolutionary consequences. Biol. Rev. 2021, 96, 433–453. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Li, W.; Hartung, J.S.; Levy, L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 2006, 66, 104–115. [Google Scholar] [CrossRef]
- Jagoueix, S.; Bove, J.-M.; Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 1994, 44, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.; Deng, X.; Hong, N.; Zhong, Y.; Wang, G.; Yi, G. Phylogenetic analysis of the citrus Huanglongbing (HLB) bacterium based on the sequences of 16S rDNA and 16S/23S rDNA intergenic regions among isolates in China. Eur. J. Plant Pathol. 2009, 124, 495–503. [Google Scholar] [CrossRef]
- Tomimura, K.; Miyata, S.I.; Furuya, N.; Kubota, K.; Okuda, M.; Subandiyah, S.; Hung, T.H.; Su, H.J.; Iwanami, T. Evaluation of genetic diversity among ‘Candidatus Liberibacter asiaticus’ isolates collected in Southeast Asia. Phytopathology 2009, 99, 1062–1069. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Flores-Cruz, Z.; Zhou, L.; Kang, B.-H.; Fleites, L.A.; Gooch, M.D.; Wulff, N.A.; Davis, M.J.; Duan, Y.-P.; Gabriel, D.W. ‘Ca. Liberibacter asiaticus’ Carries an Excision Plasmid Prophage and a Chromosomally Integrated Prophage That Becomes Lytic in Plant Infections. Mol. Plant-Microbe Interact. 2011, 24, 458–468. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Wu, F.; Zheng, Z.; Yokomi, R.; Kumagai, L.; Cai, W.; Rascoe, J.; Polek, M.; Chen, J.; Deng, X. Prophage diversity of ‘Candidatus Liberibacter asiaticus’ strains in California. Phytopathology 2019, 109, 551–559. [Google Scholar] [CrossRef]
- Zheng, Z.; Bao, M.; Wu, F.; Chen, J.; Deng, X. Predominance of Single Prophage Carrying a CRISPR/cas System in “Candidatus Liberibacter asiaticus” Strains in Southern China. PLoS ONE 2016, 11, e0146422. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, F.; Kumagai, L.Á.; Polek, M.; Deng, X.; Chen, J. Two ‘Candidatus Liberibacter asiaticus’ strains recently found in California harbor different prophages. Phytopathology 2017, 107, 662–668. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Bao, M.; Wu, F.; Van Horn, C.; Chen, J.; Deng, X. A type 3 prophage of ‘Candidatus Liberibacter asiaticus’ carrying a restriction-modification system. Phytopathology 2018, 108, 454–461. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Mirazo, M.; Jin, R.; Weitz, J.S. Functional and Comparative Genomic Analysis of Integrated Prophage-Like Sequences in “Candidatus Liberibacter asiaticus”. mSphere 2019, 4, e00409-19. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Liu, K.; Atta, S.; Zeng, C.; Zhou, C.; Wang, X. Two unique prophages of ‘Candidatus Liberibacter asiaticus’ strains from Pakistan. Phytopathology 2021, 111, 784–788. [Google Scholar] [CrossRef]
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Williams, K.P.; et al. Complete Genome Sequence of Citrus Huanglongbing Bacterium, ‘Candidatus Liberibacter asiaticus’ Obtained Through Metagenomics. Mol. Plant-Microbe Interact. 2009, 22, 1011–1020. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Deng, X.; Sun, X.; Jones, D.; Irey, M.; Civerolo, E. Guangdong and Florida Populations of ‘Candidatus Liberibacter asiaticus’ Distinguished by a Genomic Locus with Short Tandem Repeats. Phytopathology 2010, 100, 567–572. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Lopes, S.; Wang, X.; Sun, X.; Jones, D.; Irey, M.; Civerolo, E.; Chen, J. Characterization of “Candidatus Liberibacter Asiaticus” Populations by Double-Locus Analyses. Curr. Microbiol. 2014, 69, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Rascoe, J.; Kumagai, L.B.; Keremane, M.L.; Nakhla, M.K. Citrus Huanglongbing (HLB) Discoveries in California in 2015 and 2012 are of Different Genotypes of Candidatus Liberibacter asiaticus (cLas) by Double-locus Genomic Variation Analysis. Plant Dis. 2016, 100, 645. [Google Scholar] [CrossRef]
- Zhou, W.; Rousset, F.; O’Neill, S. Phylogeny and PCR–based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. B Biol. Sci. 1998, 265, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence ali gnment software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 15–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razi, M.F.; Keremane, M.L.; Ramadugu, C.; Roose, M.; Khan, I.A.; Lee, R.F. Detection of citrus Huanglongbing-associated ‘Candidatus Liberibacter asiaticus’ in citrus and Diaphorina citri in Pakistan, seasonal variability, and implications for disease management. Phytopathology 2014, 104, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Tian, S.; Xian, J.; Chen, S.; Liu, T.; Yin, Y. Detection and phylogenetic analysis of Wolbachia in the Asian citrus psyllid (Diaphorina citri) (Homoptera: Psylloidea) populations in partial areas in China. Acta Entomol. Sin. 2010, 53, 1045–1054. [Google Scholar]
- Lewis-Rosenblum, H.; Martini, X.; Tiwari, S.; Stelinski, L.L. Seasonal Movement Patterns and Long-Range Dispersal of Asian Citrus Psyllid in Florida Citrus. J. Econ. Entomol. 2015, 108, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Rao, C.N.; George, A.; Chichghare, S.A. Molecular identification and characterization of the Asian citrus psyllid vector, Diaphorina citri (Hemiptera: Psyllidae) and the transmitted Huanglongbing-associated bacterium, Candidatus Liberibacter asiaticus in India. J. Plant Pathol. 2022, 104, 1–14. [Google Scholar] [CrossRef]
- Siozios, S.; Gerth, M.; Griffin, J.S.; Hurst, G.D. Symbiosis: Wolbachia Host Shifts in the Fast Lane. Curr. Biol. 2018, 28, R269–R271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyd, E.F.; Brüsso, H. Common themes among bacteriophageencoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002, 10, 521–529. [Google Scholar] [CrossRef]
- Brüssow, H.; Canchaya, C.; Hardt, W.-D. Phages and the Evolution of Bacterial Pathogens: From Genomic Rearrangements to Lysogenic Conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleites, L.A.; Jain, M.; Zhang, S.; Gabriel, D.W. “Candidatus Liberibacter asiaticus” Prophage Late Genes May Limit Host Range and Culturability. Appl. Environ. Microbiol. 2014, 80, 6023–6030. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Fleites, L.A.; Gabriel, D.W. Prophage-encoded peroxidase in ‘Candidatus Liberibacter asiaticus’ is a secreted effector that suppresses plant defenses. Mol. Plant Microbe Interact. 2015, 28, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J., Jr. The Candidatus Liberibacter–Host Interface: Insights into Pathogenesis Mechanisms and Disease Control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef]
- Katoh, H.; Miyata, S.-I.; Inoue, H.; Iwanami, T. Unique Features of a Japanese ‘Candidatus Liberibacter asiaticus’ Strain Revealed by Whole Genome Sequencing. PLoS ONE 2014, 9, e106109. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Sun, X.; Deng, X.; Chen, J. Whole-Genome Sequence of “Candidatus Liberibacter asiaticus” from a Huanglongbing-Affected Citrus Tree in Central Florida. Genome Announc. 2015, 3, e00169-15. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, Y.E.; Paredes-Montero, J.R.; Al-Saleh, M.A.; Widyawan, A.; He, R.; El Komy, M.H.; Al Dhafer, H.M.; Kitchen, N.; Gang, D.R.; Brown, J.K. Characterization of the Asian Citrus Psyllid-‘Candidatus Liberibacter Asiaticus’ Pathosystem in Saudi Arabia Reveals Two Predominant CLas Lineages and One Asian Citrus Psyllid Vector Haplotype. Microorganisms 2022, 10, 1991. https://doi.org/10.3390/microorganisms10101991
Ibrahim YE, Paredes-Montero JR, Al-Saleh MA, Widyawan A, He R, El Komy MH, Al Dhafer HM, Kitchen N, Gang DR, Brown JK. Characterization of the Asian Citrus Psyllid-‘Candidatus Liberibacter Asiaticus’ Pathosystem in Saudi Arabia Reveals Two Predominant CLas Lineages and One Asian Citrus Psyllid Vector Haplotype. Microorganisms. 2022; 10(10):1991. https://doi.org/10.3390/microorganisms10101991
Chicago/Turabian StyleIbrahim, Yasser E., Jorge R. Paredes-Montero, Mohammed A. Al-Saleh, Arya Widyawan, Ruifeng He, Mahmoud H. El Komy, Hathal M. Al Dhafer, Noel Kitchen, David R. Gang, and Judith K. Brown. 2022. "Characterization of the Asian Citrus Psyllid-‘Candidatus Liberibacter Asiaticus’ Pathosystem in Saudi Arabia Reveals Two Predominant CLas Lineages and One Asian Citrus Psyllid Vector Haplotype" Microorganisms 10, no. 10: 1991. https://doi.org/10.3390/microorganisms10101991




