Longitudinal Study on Extended-Spectrum Beta-Lactamase-E. coli in Sentinel Mallard Ducks in an Important Baltic Stop-Over Site for Migratory Ducks in Germany
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Ethical Approval
2.3. Bacterial Isolation
2.4. Antimicrobial Susceptibility Testing (AST) Using VITEK 2
2.5. Whole-Genome Sequencing (WGS) and Analysis
3. Results
3.1. Bacterial Isolation
3.2. Antimicrobial Susceptibility Testing (AST)
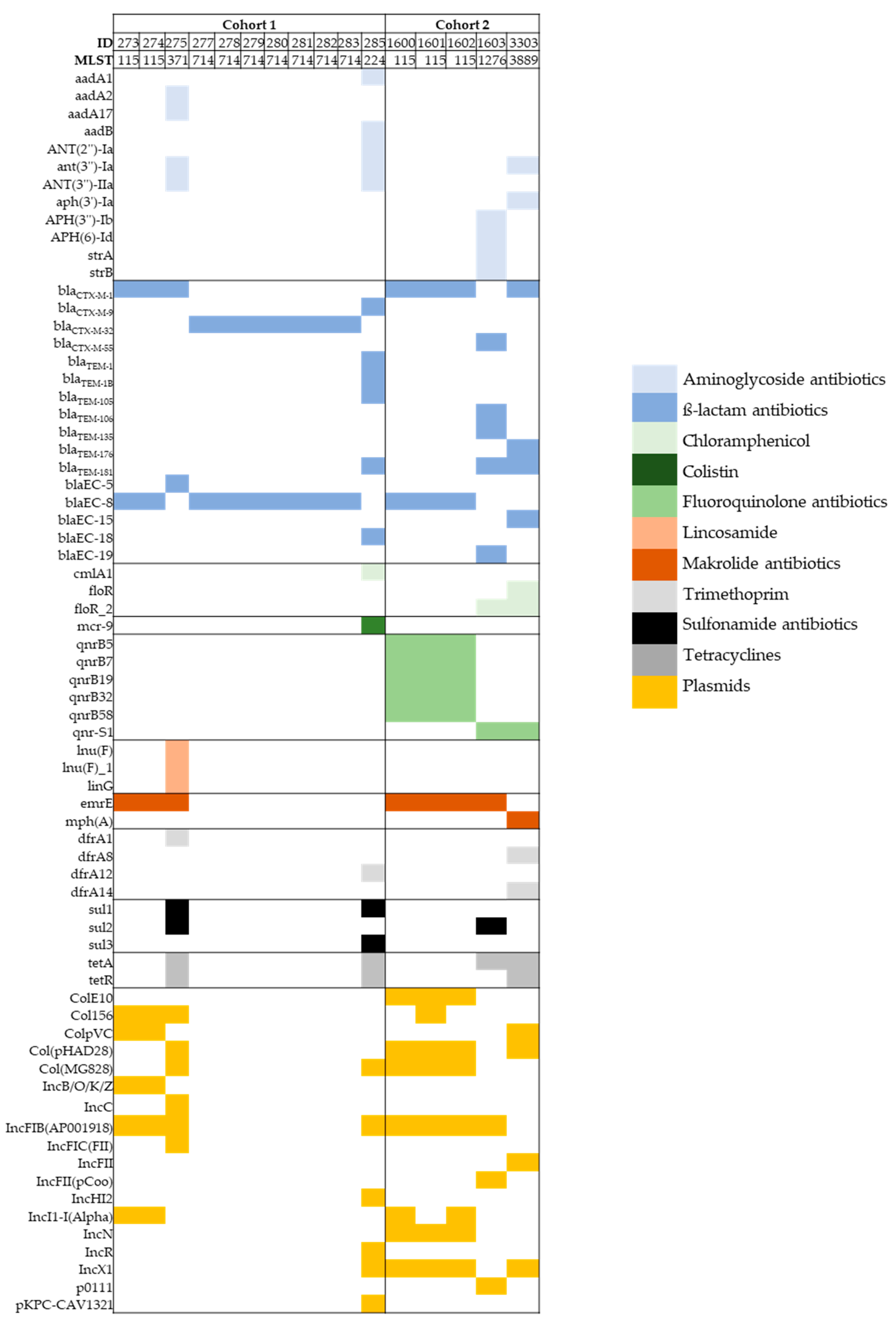
3.3. Whole-Genome Sequence (WGS) Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.F.; Diez, M.J.; Sahagun, A.M.; Diez, R.; Sierra, M.; Garcia, J.J.J.; Lopez, C.; Fernandez, M.N. Availability of Antibiotics for Veterinary Use on the Internet: A Cross-Sectional Study. Front. Vet. Sci. 2022, 8, 1673. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef]
- ECDC. Factsheet for Experts-Antimicrobial Resistance. Available online: https://www.ecdc.europa.eu/en/antimicrobial-resistance/facts/factsheets/experts (accessed on 30 August 2022).
- Homeier-Bachmann, T.; Heiden, S.E.; Lubcke, P.K.; Bachmann, L.; Bohnert, J.A.; Zimmermann, D.; Schaufler, K. Antibiotic-Resistant Enterobacteriaceae in Wastewater of Abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Zheng, Y.C.; Yu, S.; Wang, G.Q.; Xie, F.C.; Xu, H.F.; Du, S.D.; Zhao, H.T.; Sang, X.T.; Lu, J.Z.; Jiang, W.J. Comparative microbial antibiotic resistome between urban and deep forest environments. Environ. Microbiol. Rep. 2021, 13, 503–508. [Google Scholar] [CrossRef]
- McCann, C.M.; Christgen, B.; Roberts, J.A.; Su, J.Q.; Arnold, K.E.; Gray, N.D.; Zhu, Y.G.; Graham, D.W. Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 2019, 125, 497–504. [Google Scholar] [CrossRef]
- Marinho, C.; Silva, N.; Pombo, S.; Santos, T.; Monteiro, R.; Goncalves, A.; Micael, J.; Rodrigues, P.; Costa, A.C.; Igrejas, G.; et al. Echinoderms from Azores islands: An unexpected source of antibiotic resistant Enterococcus spp. and Escherichia coli isolates. Mar. Pollut. Bull. 2013, 69, 122–127. [Google Scholar] [CrossRef]
- Homeier-Bachmann, T.; Schuetz, A.K.; Dreyer, S.; Glanz, J.; Schaufler, K.; Conraths, F.J. Genomic Analysis of ESBL-Producing E. coli in Wildlife from North-Eastern Germany. Antibiotics 2022, 11, 123. [Google Scholar] [CrossRef]
- Plaza-Rodriguez, C.; Alt, K.; Grobbel, M.; Hammerl, J.A.; Irrgang, A.; Szabo, I.; Stingl, K.; Schuh, E.; Wiehle, L.; Pfefferkorn, B.; et al. Wildlife as Sentinels of Antimicrobial Resistance in Germany? Front. Vet. Sci. 2021, 7, 627821. [Google Scholar] [CrossRef]
- Wyrsch, E.R.; Nesporova, K.; Tarabai, H.; Jamborova, I.; Bitar, I.; Literak, I.; Dolejska, M.; Djordjevic, S.P. Urban Wildlife Crisis: Australian Silver Gull Is a Bystander Host to Widespread Clinical Antibiotic Resistance. Msystems 2022, 7, e00158-22. [Google Scholar] [CrossRef]
- Khan, S.A.; Imtiaz, M.A.; Sayeed, M.A.; Shaikat, A.H.; Hassan, M.M. Antimicrobial resistance pattern in domestic animal-wildlife-environmental niche via the food chain to humans with a Bangladesh perspective; a systematic review. BMC Vet. Res. 2020, 16, 302. [Google Scholar] [CrossRef]
- Grunzweil, O.M.; Palmer, L.; Cabal, A.; Szostak, M.P.; Ruppitsch, W.; Kornschober, C.; Korus, M.; Misic, D.; Bernreiter-Hofer, T.; Korath, A.D.J.; et al. Presence of beta-Lactamase-producing Enterobacterales and Salmonella Isolates in Marine Mammals. Int. J. Mol. Sci. 2021, 22, 5905. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, I.N.; Fernandez-Fernandez, R.; Juarez-Fernandez, G.; Martinez-Alvarez, S.; Eguizabal, P.; Zarazaga, M.; Lozano, C.; Torres, C. Wild Animals Are Reservoirs and Sentinels of Staphylococcus aureus and MRSA Clones: A Problem with “One Health” Concern. Antibiotics 2021, 10, 1556. [Google Scholar] [CrossRef]
- Melendez, D.; Roberts, M.C.; Greninger, A.L.; Weissman, S.; No, D.; Rabinowitz, P.; Wasser, S. Whole-genome analysis of extraintestinal pathogenic Escherichia coli (ExPEC) MDR ST73 and ST127 isolated from endangered southern resident killer whales (Orcinus orca). J. Antimicrob. Chemother. 2019, 74, 2176–2180. [Google Scholar] [CrossRef]
- Mora, A.; Garcia-Pena, F.J.; Alonso, M.P.; Pedraza-Diaz, S.; Ortega-Mora, L.M.; Garcia-Parraga, D.; Lopez, C.; Viso, S.; Dahbi, G.; Marzoa, J.; et al. Impact of human-associated Escherichia coli clonal groups in Antarctic pinnipeds: Presence of ST73, ST95, ST141 and ST131. Sci. Rep. 2018, 8, 4678. [Google Scholar] [CrossRef] [Green Version]
- Nguema, P.P.M.; Onanga, R.; Atome, G.R.N.; Mbeang, J.C.O.; Mabika, A.M.; Yaro, M.; Lounnas, M.; Dumont, Y.; Zohra, Z.F.; Godreuil, S.; et al. Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon. Microorganisms 2020, 8, 138. [Google Scholar] [CrossRef] [Green Version]
- Vasquez-Aguilar, A.A.; Toledo-Manuel, F.O.; Barbachano-Guerrero, A.; Hernandez-Rodriguez, D. Detection of Antimicrobial Resistance Genes in Escherichia coli Isolated from Black Howler Monkeys (Alouatta pigra) and Domestic Animals in Fragmented Rain-Forest Areas in Tabasco, Mexico. J. Wildl. Dis. 2020, 56, 922–927. [Google Scholar] [CrossRef]
- Nowak, K.; Fahr, J.; Weber, N.; Lubke-Becker, A.; Semmler, T.; Weiss, S.; Mombouli, J.V.; Wieler, L.H.; Guenther, S.; Leendertz, F.H.; et al. Highly diverse and antimicrobial susceptible Escherichia coli display a naive bacterial population in fruit bats from the Republic of Congo. PLoS ONE 2017, 12, e0178146. [Google Scholar] [CrossRef] [Green Version]
- Formenti, N.; Calo, S.; Parisio, G.; Guarneri, F.; Birbes, L.; Pitozzi, A.; Scali, F.; Tonni, M.; Guadagno, F.; Giovannini, S.; et al. ESBL/AmpC-Producing Escherichia coli in Wild Boar: Epidemiology and Risk Factors. Animals 2021, 11, 1855. [Google Scholar] [CrossRef]
- Schierack, P.; Heiden, S.E.; Khan, M.M.; Nikolaus, L.; Kolenda, R.; Stubbe, M.; Lkhagvasuren, D.; Rodiger, S.; Guenther, S.; Schaufler, K. Genomic and Phenotypic Analysis of an ESBL-ProducingE. coliST1159 Clonal Lineage From Wild Birds in Mongolia. Front. Microbiol. 2020, 11, 1699. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Semmler, T.; Stubbe, A.; Stubbe, M.; Wieler, L.H.; Schaufler, K. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J. Antimicrob. Chemother. 2017, 72, 1310–1313. [Google Scholar] [CrossRef] [PubMed]
- Van den Honert, M.S.; Gouws, P.A.; Hoffman, L.C. Escherichia coli Antibiotic Resistance Patterns from Co-Grazing and Non-Co-Grazing Livestock and Wildlife Species from Two Farms in the Western Cape, South Africa. Antibiotics 2021, 10, 618. [Google Scholar] [CrossRef]
- Lundback, I.C.; McDougall, F.K.; Dann, P.; Slip, D.J.; Gray, R.; Power, M.L. Into the sea: Antimicrobial resistance determinants in the microbiota of little penguins (Eudyptula minor). Infect. Genet. Evol. 2021, 88, 104697. [Google Scholar] [CrossRef] [PubMed]
- Jara, D.; Bello-Toledo, H.; Dominguez, M.; Cigarroa, C.; Fernandez, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; Gonzalez-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramey, A.M.; Hernandez, J.; Tyrlov, V.; Uher-Koch, B.D.; Schmutz, J.A.; Atterby, C.; Jarhult, J.D.; Bonnedahl, J. Antibiotic-Resistant Escherichia coli in Migratory Birds Inhabiting Remote Alaska. EcoHealth 2018, 15, 72–81. [Google Scholar] [CrossRef]
- Lorenti, E.; Moredo, F.; Origlia, J.; Diaz, J.I.; Cremonte, F.; Giacoboni, G. Gulls as carriers of antimicrobial resistance genes in different biogeographical areas of South America. An. Acad. Bras. Cienc. 2021, 93. [Google Scholar] [CrossRef]
- Mohsin, M.; Raza, S.; Schaufler, K.; Roschanski, N.; Sarwar, F.; Semmler, T.; Schierack, P.; Guenther, S. High Prevalence of CTX-M-15-Type ESBL-Producing E. coil from Migratory Avian Species in Pakistan. Front. Microbiol. 2017, 8, 2476. [Google Scholar] [CrossRef] [Green Version]
- Fashae, K.; Engelmann, I.; Monecke, S.; Braun, S.D.; Ehricht, R. Molecular characterisation of extended-spectrum ß-lactamase producing Escherichia coli in wild birds and cattle, Ibadan, Nigeria. BMC Vet. Res. 2021, 17, 33. [Google Scholar] [CrossRef]
- Davies, Y.M.; Cunha, M.P.V.; Oliveira, M.G.X.; Oliveira, M.C.V.; Philadelpho, N.; Romero, D.C.; Milanelo, L.; Guimaraes, M.B.; Ferreira, A.J.P.; Moreno, A.M.; et al. Virulence and antimicrobial resistance of Klebsiella pneumoniae isolated from passerine and psittacine birds. Avian Pathol. 2016, 45, 194–201. [Google Scholar] [CrossRef]
- Torres, R.T.; Fernandes, J.; Carvalho, J.; Cunha, M.V.; Caetano, T.; Mendo, S.; Serrano, E.; Fonseca, C. Wild boar as a reservoir of antimicrobial resistance. Sci. Total Environ. 2020, 717, 135001. [Google Scholar] [CrossRef] [PubMed]
- Furness, L.E.; Campbell, A.; Zhang, L.H.; Gaze, W.H.; McDonald, R.A. Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ. Res. 2017, 154, 28–34. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of beta-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef]
- Globig, A.; Baumer, A.; Revilla-Fernández, S.; Beer, M.; Wodak, E.; Fink, M.; Greber, N.; Harder, T.C.; Wilking, H.; Brunhart, I.; et al. Ducks as sentinels for avian influenza in wild birds. Emerg. Infect. Dis. 2009, 15, 1633–1636. [Google Scholar] [CrossRef]
- Koethe, S.; Ulrich, L.; Ulrich, R.; Amler, S.; Graaf, A.; Harder, T.C.; Grund, C.; Mettenleiter, T.C.; Conraths, F.J.; Beer, M.; et al. Modulation of lethal HPAIV H5N8 clade 2.3.4.4B infection in AIV pre-exposed mallards. Emerg. Microbes Infect. 2020, 9, 180–193. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.G.; Bean, D.C.; Clarke, R.H.; Loyn, R.; Larkins, J.A.; Hassell, C.; Greenhill, A.R. Presence and antimicrobial resistance profiles of Escherichia coli, Enterococcusspp. and Salmonellasp. in 12 species of Australian shorebirds and terns. Zoonoses Public Health 2022, 69, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, C.A.; Woksepp, H.; Sandegren, L.; Mohsin, M.; Hasan, B.; Muzyka, D.; Hernandez, J.; Aguirre, F.; Tok, A.; Soderman, J.; et al. Genomically diverse carbapenem resistant Enterobacteriaceae fromwild birds provide insight into global patterns of spatiotemporal dissemination. Sci. Total Environ. 2022, 824, 153632. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Z.; M’Ikanatha, N.M.; Dudley, E.G. Comparative Genomic Analysis of Salmonella enterica Serovar Typhimurium Isolates from Passerines Reveals Two Lineages Circulating in Europe, New Zealand, and the United States. Appl. Environ. Microbiol. 2022, 88, e00205-22. [Google Scholar] [CrossRef] [PubMed]
- Plawinska-Czarnak, J.; Wodz, K.; Piechowicz, L.; Tokarska-Pietrzak, E.; Belkot, Z.; Bogdan, J.; Wisniewski, J.; Kwiecinski, P.; Kwiecinski, A.; Anusz, K. Wild Duck (Anas platyrhynchos) as a Source of Antibiotic-Resistant Salmonella enterica subsp. diarizonae O58-The First Report in Poland. Antibiotics 2022, 11, 530. [Google Scholar] [CrossRef]
- Retamal, P.; Llanos-Soto, S.; Salas, L.M.; Lopez, J.; Vianna, J.; Hernandez, J.; Medina-Vogel, G.; Castaneda, F.; Fresno, M.; Gonzalez-Acuna, D. Isolation of drug-resistant Salmonella enterica serovar enteritidis strains in gentoo penguins from Antarctica. Polar Biol. 2017, 40, 2531–2536. [Google Scholar] [CrossRef]
- Ong, K.H.; Khor, W.C.; Quek, J.Y.; Low, Z.X.; Arivalan, S.; Humaidi, M.; Chua, C.; Seow, K.L.G.; Guo, S.; Tay, M.Y.F.; et al. Occurrence and Antimicrobial Resistance Traits of Escherichia coli from Wild Birds and Rodents in Singapore. Int. J. Environ. Res. Public Health 2020, 17, 5606. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Imirzalioglu, C.; Oppong, K.; Akenten, C.W.; Hogan, B.; Krumkamp, R.; Poppert, S.; Levermann, V.; Schwengers, O.; Sarpong, N.; et al. Detection and Characterization of ESBL-Producing Escherichia coli From Humans and Poultry in Ghana. Front. Microbiol. 2018, 9, 3358. [Google Scholar] [CrossRef]
- Badr, H.; Reda, R.M.; Hagag, N.M.; Kamel, E.; Elnomrosy, S.M.; Mansour, A.I.; Shahein, M.A.; Ali, S.F.; Ali, H.R. Multidrug-Resistant and Genetic Characterization of Extended-Spectrum Beta-Lactamase-Producing E. coli Recovered from Chickens and Humans in Egypt. Animals 2022, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Vounba, P.; Arsenault, J.; Bada-Alambedji, R.; Fairbrother, J.M. Pathogenic potential and the role of clones and plasmids in beta-lactamase-producing E. coli from chicken faeces in Vietnam. BMC Vet. Res. 2019, 15, 106. [Google Scholar] [CrossRef] [PubMed]
- Bastard, J.; Nhung, N.T.; Hien, V.B.; Kiet, B.T.; Temime, L.; Opatowski, L.; Carrique-Mas, J.; Choisy, M. Modelling the impact of antimicrobial use and external introductions on commensal E. coli colistin resistance in small-scale chicken farms of the Mekong delta of Vietnam. Transbound. Emerg. Dis. 2022, 69, e2185–e2194. [Google Scholar] [CrossRef] [PubMed]
- Rabbia, V.; Bello-Toledo, H.; Jimenez, S.; Quezada, M.; Dominguez, M.; Vergara, L.; Gomez-Fuentes, C.; Calisto-Ulloa, N.; Gonzalez-Acuna, D.; Lopez, J.; et al. Antibiotic resistance in Escherichia coli strains isolated from Antarctic bird feces, water from inside a wastewater treatment plant, and seawater samples collected in the Antarctic Treaty area. Polar Sci. 2016, 10, 123–131. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Bendjama, E.; Cherak, Z.; Khellaf, M.; Khemri, A.; Rolain, J.M. Detection of bla(OXA-48) and mcr-1 Genes in Escherichia coli Isolates from Pigeon (Columba livia) in Algeria. Microorganisms 2022, 10, 975. [Google Scholar] [CrossRef]
- Abreu, R.; Castro, B.; Espigares, E.; Rodriguez-Alvarez, C.; Lecuona, M.; Moreno, E.; Espigares, M.; Arias, A. Prevalence of CTX-M-Type Extended-Spectrum beta-Lactamases in Escherichia coli Strains Isolated in Poultry Farms. Foodborne Pathog. Dis. 2014, 11, 868–873. [Google Scholar] [CrossRef]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e0002521. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.D.E.; Canica, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum beta-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Zhou, Z.M.; Alikhan, N.F.; Mohamed, K.; Fan, Y.L.; Achtman, M.; Brown, D.; Chattaway, M.; Dallman, T.; Delahay, R.; Kornschober, C.; et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Villa, L.; Fortini, D.; García-Fernández, A. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 2021, 118, 102392. [Google Scholar] [CrossRef]
- Skarzynska, M.; Zajac, M.; Bomba, A.; Bocian, L.; Kozdrun, W.; Polak, M.; Wiacek, J.; Wasyl, D. Antimicrobial Resistance Glides in the Sky-Free-Living Birds as a Reservoir of Resistant Escherichia coli With Zoonotic Potential. Front. Microbiol. 2021, 12, 656223. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.L.; Zhang, Z.F.; Shen, H.; Ning, M.Z.; Chen, J.H.; Wei, H.X.; Zhang, K. Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. Apmis 2014, 122, 1088–1095. [Google Scholar] [CrossRef]
- Ramadan, H.; Jackson, C.R.; Frye, J.G.; Hiott, L.M.; Samir, M.; Awad, A.; Woodley, T.A. Antimicrobial Resistance, Genetic Diversity and Multilocus Sequence Typing of Escherichia coli from Humans, Retail Chicken and Ground Beef in Egypt. Pathogens 2020, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-spectrum β-lactamase-producing Enterobacteriaceae from animal origin and wastewater in Tunisia: First detection of O25b-B2(3)-CTX-M-27-ST131 Escherichia coli and CTX-M-15/OXA-204-producing Citrobacter freundii from wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M.; Turner, M.S.; Siragusa, G.; White, D. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [Green Version]
- Martiny, H.M.; Munk, P.; Brinch, C.; Szarvas, J.; Aarestrup, F.M.; Petersen, T.N. Global Distribution of mcr Gene Variants in 214K Metagenomic Samples. Msystems 2022, 7, e00105-22. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Li, C.; Hsu, C.H.; Ayers, S.; Borenstein, S.; Mukherjee, S.; Tran, T.T.; McDermott, P.F.; Zhao, S. The mcr-9 Gene of Salmonella and Escherichia coli Is Not Associated with Colistin Resistance in the United States. Antimicrob. Agents Chemother. 2020, 64, e00573-20. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Royer, G.; Decousser, J.W.; Bourrel, A.S.; Palmieri, M.; De La Rosa, J.M.O.; Jacquier, H.; Denamur, E.; Nordmann, P.; Poirel, L. mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 2019, 63, e00965-19. [Google Scholar] [CrossRef] [Green Version]
- Chaalal, N. Colistin-Resistant Enterobacterales Isolated from Chicken Meat in Western Algeria. Microb. Drug Resist. 2021, 27, 991–1002. [Google Scholar] [CrossRef]
- Yamasaki, S.; Le, T.D.; Vien, M.Q.; Van Dang, C.; Yamamoto, Y. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and residual antimicrobials in the environment in Vietnam. Anim. Health Res. Rev. 2017, 18, 128–135. [Google Scholar] [CrossRef]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitout, J.D.D.; Peirano, G.; Chen, L.; DeVinney, R.; Matsumura, Y. Escherichia coli ST1193: Following in the Footsteps of E. coli ST131. Antimicrob. Agents Chemother. 2022, 66, e00511-22. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, M.; Igarashi, M.; Watanabe, H.; Qin, L.; Ohnishi, M.; Terajima, J.; Iyoda, S.; Morita-Ishihara, T.; Tateda, K.; Ishii, Y.; et al. Epidemiology and genotypic characterisation of dissemination patterns of uropathogenic Escherichia coli in a community. Epidemiol. Infect. 2019, 147, e148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, F.B.; Nielsen, J.B.; Schønning, K.; Littauer, P.; Knudsen, J.D.; Løbner-Olesen, A.; Frimodt-Møller, N. “Population structure of drug-susceptible,-resistant and ESBL-producing Escherichia coli from community-acquired urinary tract”. BMC Microbiol. 2016, 16, 63. [Google Scholar] [CrossRef] [Green Version]
- Haenni, M.; Métayer, V.; Jarry, R.; Drapeau, A.; Puech, M.P.; Madec, J.Y.; Keck, N. Wide Spread of bla (CTX-M-9)/mcr-9 IncHI2/ST1 Plasmids and CTX-M-9-Producing Escherichia coli and Enterobacter cloacae in Rescued Wild Animals. Front. Microbiol. 2020, 11, 601317. [Google Scholar] [CrossRef]
- Eger, E.; Domke, M.; Heiden, S.E.; Paditz, M.; Balau, V.; Huxdorff, C.; Zimmermann, D.; Homeier-Bachmann, T.; Schaufler, K. Highly Virulent and Multidrug-Resistant Escherichia coli Sequence Type 58 from a Sausage in Germany. Antibiotics 2022, 11, 1006. [Google Scholar] [CrossRef]
- Kluytmans-van den Bergh, M.F.Q.; Rossen, J.W.A.; Bruijning-Verhagen, P.C.J.; Bonten, M.J.M.; Friedrich, A.W.; Vandenbroucke-Grauls, C.; Willems, R.J.L.; Kluytmans, J.; So, M.S.G. Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 2919–2927. [Google Scholar] [CrossRef] [Green Version]
- Tahar, S.; Nabil, M.M.; Safia, T.; Ngaiganam, E.P.; Omar, A.; Hafidha, C.; Hanane, Z.; Rolain, J.M.; Diene, S.M. Molecular Characterization of Multidrug-Resistant Escherichia coli Isolated from Milk of Dairy Cows with Clinical Mastitis in Algeria. J. Food Prot. 2020, 83, 2173–2178. [Google Scholar] [CrossRef]
- Olsen, R.H.; Bisgaard, M.; Löhren, U.; Robineau, B.; Christensen, H. Extended-spectrum β-lactamase-producing Escherichia coli isolated from poultry: A review of current problems, illustrated with some laboratory findings. Avian Pathol. 2014, 43, 199–208. [Google Scholar] [CrossRef]
- Poulsen, L.L.; Bisgaard, M.; Jørgensen, S.L.; Dideriksen, T.; Pedersen, J.R.; Christensen, H. Characterization of Escherichia coli causing cellulitis in broilers. Vet. Microbiol. 2018, 225, 72–78. [Google Scholar] [CrossRef]
- Randall, L.P.; Clouting, C.; Horton, R.A.; Coldham, N.G.; Wu, G.; Clifton-Hadley, F.A.; Davies, R.H.; Teale, C.J. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 2011, 66, 86–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, C.; Troxler, S.; Jandreski-Cvetkovic, D.; Zloch, A.; Hess, M. Escherichia coli Isolated from Organic Laying Hens Reveal a High Level of Antimicrobial Resistance despite No Antimicrobial Treatments. Antibiotics 2022, 11, 467. [Google Scholar] [CrossRef] [PubMed]
- Landman, W.J.M.; Buter, G.J.; Dijkman, R.; van Eck, J.H.H. In vivo typing of Escherichia coli obtained from laying chickens with the E. coli peritonitis syndrome. Avian Pathol. 2021, 50, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; van Hoek, A.H.; Hamidjaja, R.A.; van der Plaats, R.Q.; Kerkhof-de Heer, L.; de Roda Husman, A.M.; Schets, F.M. Distribution, Numbers, and Diversity of ESBL-Producing E. coli in the Poultry Farm Environment. PLoS ONE 2015, 10, e0135402. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhu, T.; Zhou, D.; Lu, W.; Liu, H.; Sun, Z.; Ying, J.; Lu, J.; Lin, X.; Li, K.; et al. Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell Infect. Microbiol. 2020, 10, 369. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Kang, H.S.; Kim, Y.; Kim, M.; Kwak, H.; Ryu, S. Prevalence and Genetic Characterization of mcr-1-Positive Escherichia coli Isolated from Retail Meats in South Korea. J. Microbiol. Biotechnol. 2020, 30, 1862–1869. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Li, X.; Ma, L.; Cao, X.; Hu, W.; Zhao, L.; Jing, W.; Lan, X.; Li, Y.; et al. Genetic diversity, antimicrobial resistance and extended-spectrum β-lactamase type of Escherichia coli isolates from chicken, dog, pig and yak in Gansu and Qinghai Provinces, China. J. Glob. Antimicrob. Resist. 2020, 22, 726–732. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]

| Cohort | Year | Sampling Date | Positives/Number of Samples Taken | Sample Type | Isolate No. (Origin of Sample) |
|---|---|---|---|---|---|
| 1 | 2020 | 27 May | 1/15 | C | 274 (11) |
| 10 June | 1/15 | C | 273 (5) | ||
| 1 | F | 275 (pooled, non-duck) | |||
| 25 June | 1/15 | C | 285 (5) | ||
| 8 July | 6/15 | C | 277–282 (1–3, 9, 10, 13) | ||
| 1 | F | 283 (pooled) | |||
| 22 July | 0/15 | C | |||
| 5 August | 0/15 | C | |||
| 18 August | 0/15 | C | |||
| 1 September | 0/15 | C | |||
| 16 September | 0/15 | C | |||
| 30 September | 0/15 | C | |||
| 13 October | 0/15 | C | |||
| 11 November | 0/15 | C | |||
| 2 December | 0/15 | C | |||
| 22 December | 0/15 | C | |||
| 2021 | 19 January | 0/15 | C | ||
| 2 | 2021 | 3 June | 3/10 | C | 1600–1602 (1, 2, 10) |
| 1 | F | 1603 * (pooled) | |||
| 17 June | 0/10 | C | |||
| 8 July | 0/9 | C | |||
| 30 July | 0/9 | C | |||
| 26 August | 0/10 | C | |||
| 14 September | 0/12 | C | |||
| 26 October | 0/7 | C | |||
| 8/9 November | 0/5 | C | |||
| 17 November | 0/7 | C | |||
| 23 November | 0/6 | C | |||
| 15 December | 0/8 | C | |||
| 2022 | 14 January | 0/5 | C | ||
| 3 February | 0/7 | C | |||
| 24 February | 0/8 | C | |||
| 9 March | 0/4 | C | |||
| March | 1/13 | C | 3303 (12) |
| Cohort | Designation | AMO | AMP | AMO/CA | CFL | CTX | CAZ | CFZ/TAZ | FEP | IMI | MEM | AMI | GEN | TOB | CIP | TGC | FOS | COL | TMP/SMX | ESBL | MDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 273 | X | |||||||||||||||||||
| 274 | X | ||||||||||||||||||||
| 275 | X | X | |||||||||||||||||||
| 277 | X | ||||||||||||||||||||
| 278 | X | ||||||||||||||||||||
| 279 | X | ||||||||||||||||||||
| 280 | X | ||||||||||||||||||||
| 281 | X | ||||||||||||||||||||
| 282 | X | ||||||||||||||||||||
| 283 | X | ||||||||||||||||||||
| 285 | X | X | |||||||||||||||||||
| 2 | 1600 | X | |||||||||||||||||||
| 1601 | X | ||||||||||||||||||||
| 1602 | X | ||||||||||||||||||||
| 1603 | X | ||||||||||||||||||||
| 3303 | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dreyer, S.; Globig, A.; Bachmann, L.; Schütz, A.K.; Schaufler, K.; Homeier-Bachmann, T. Longitudinal Study on Extended-Spectrum Beta-Lactamase-E. coli in Sentinel Mallard Ducks in an Important Baltic Stop-Over Site for Migratory Ducks in Germany. Microorganisms 2022, 10, 1968. https://doi.org/10.3390/microorganisms10101968
Dreyer S, Globig A, Bachmann L, Schütz AK, Schaufler K, Homeier-Bachmann T. Longitudinal Study on Extended-Spectrum Beta-Lactamase-E. coli in Sentinel Mallard Ducks in an Important Baltic Stop-Over Site for Migratory Ducks in Germany. Microorganisms. 2022; 10(10):1968. https://doi.org/10.3390/microorganisms10101968
Chicago/Turabian StyleDreyer, Sylvia, Anja Globig, Lisa Bachmann, Anne K. Schütz, Katharina Schaufler, and Timo Homeier-Bachmann. 2022. "Longitudinal Study on Extended-Spectrum Beta-Lactamase-E. coli in Sentinel Mallard Ducks in an Important Baltic Stop-Over Site for Migratory Ducks in Germany" Microorganisms 10, no. 10: 1968. https://doi.org/10.3390/microorganisms10101968
APA StyleDreyer, S., Globig, A., Bachmann, L., Schütz, A. K., Schaufler, K., & Homeier-Bachmann, T. (2022). Longitudinal Study on Extended-Spectrum Beta-Lactamase-E. coli in Sentinel Mallard Ducks in an Important Baltic Stop-Over Site for Migratory Ducks in Germany. Microorganisms, 10(10), 1968. https://doi.org/10.3390/microorganisms10101968






