Measuring Escherichia coli Gene Expression during Human Urinary Tract Infections
Abstract
:1. Introduction
2. Importance of Measuring Bacterial Gene Expression in the Host
3. Lessons from E. coli Help Redefine Bacterial Virulence
4. The ExPEC Pathotype Helps Make the Case
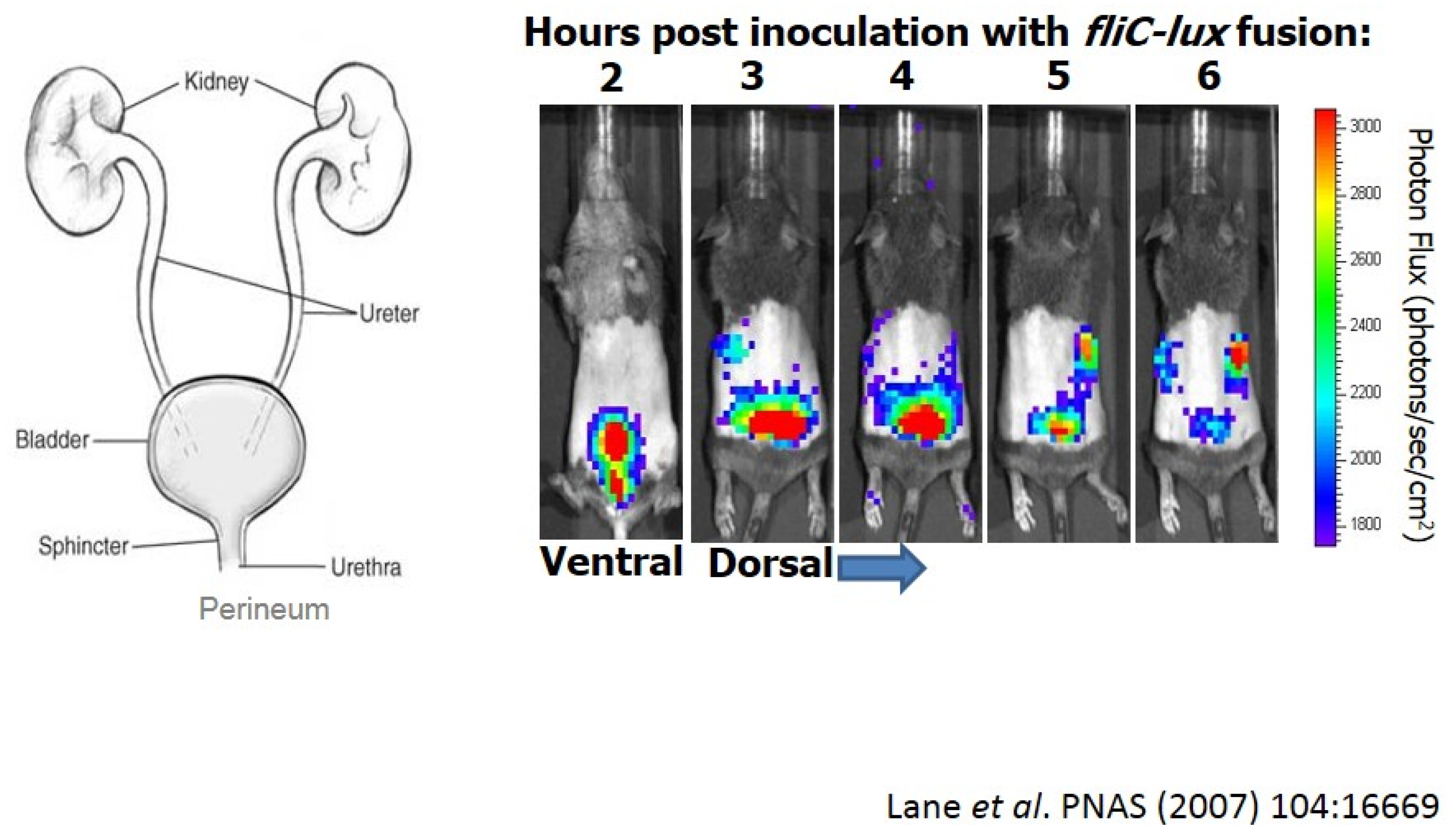
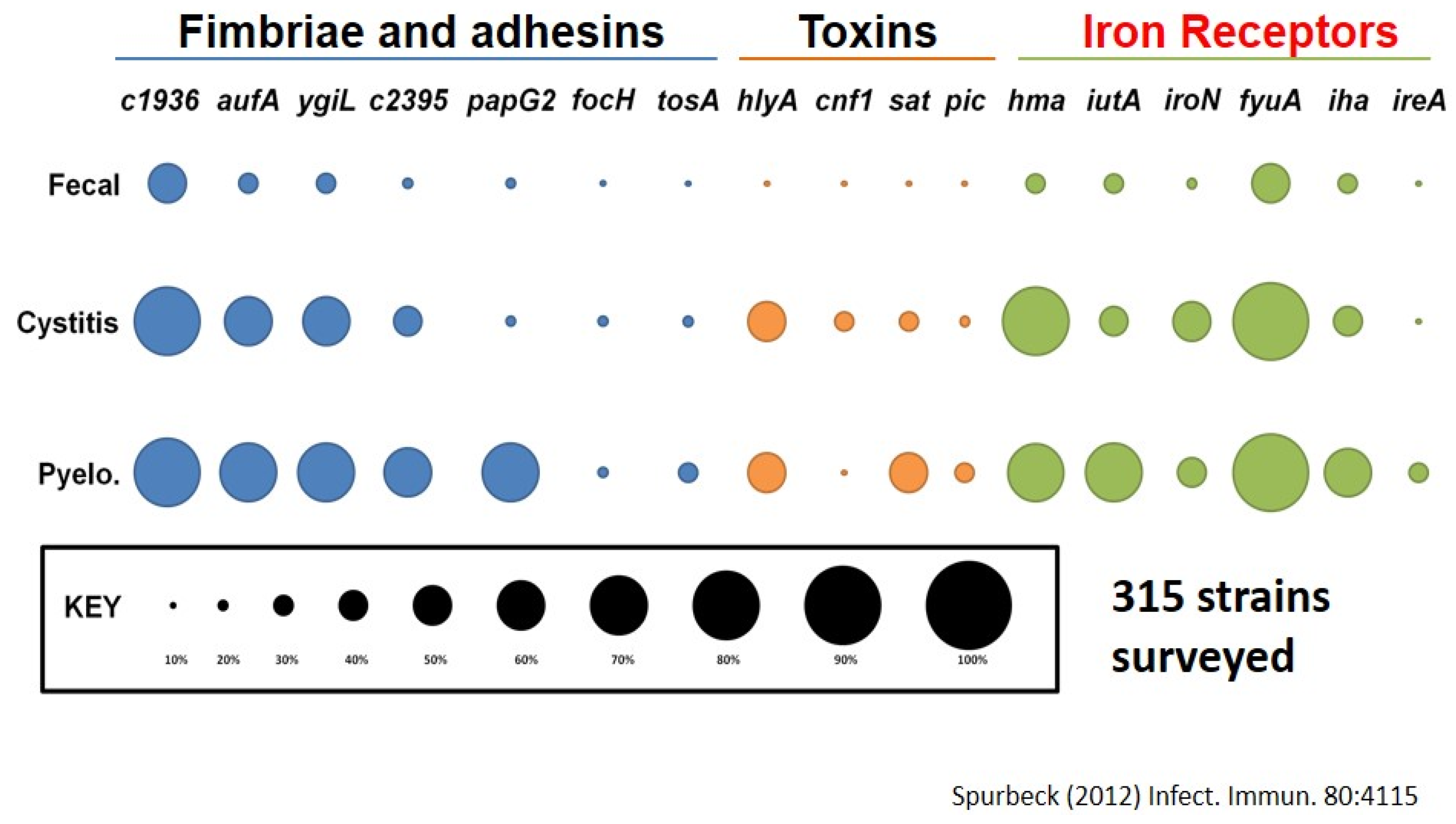
5. What Else Should We Consider to Define Bacterial Virulence?
6. Measuring Global Gene Expression during Bacterial Infection
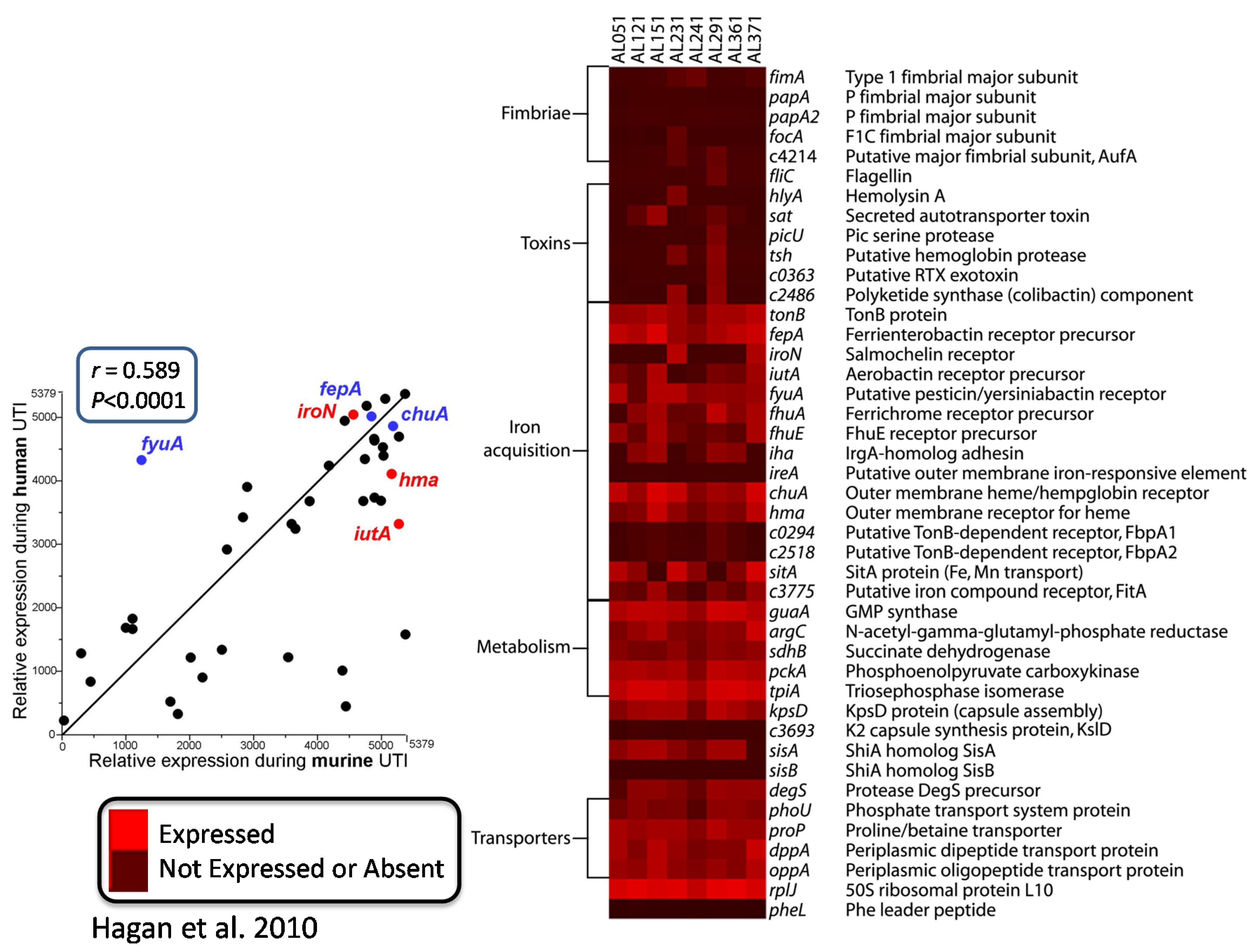
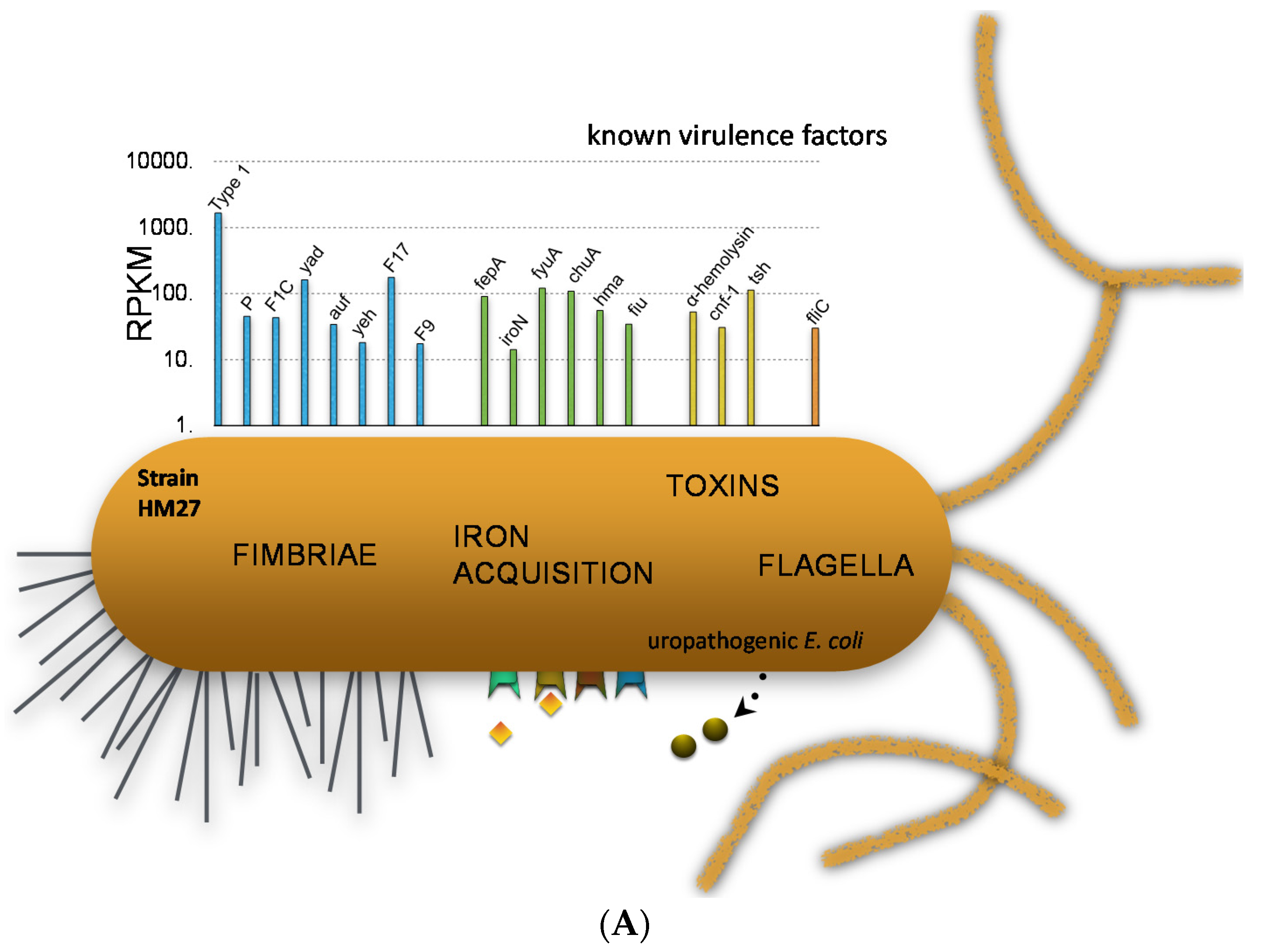
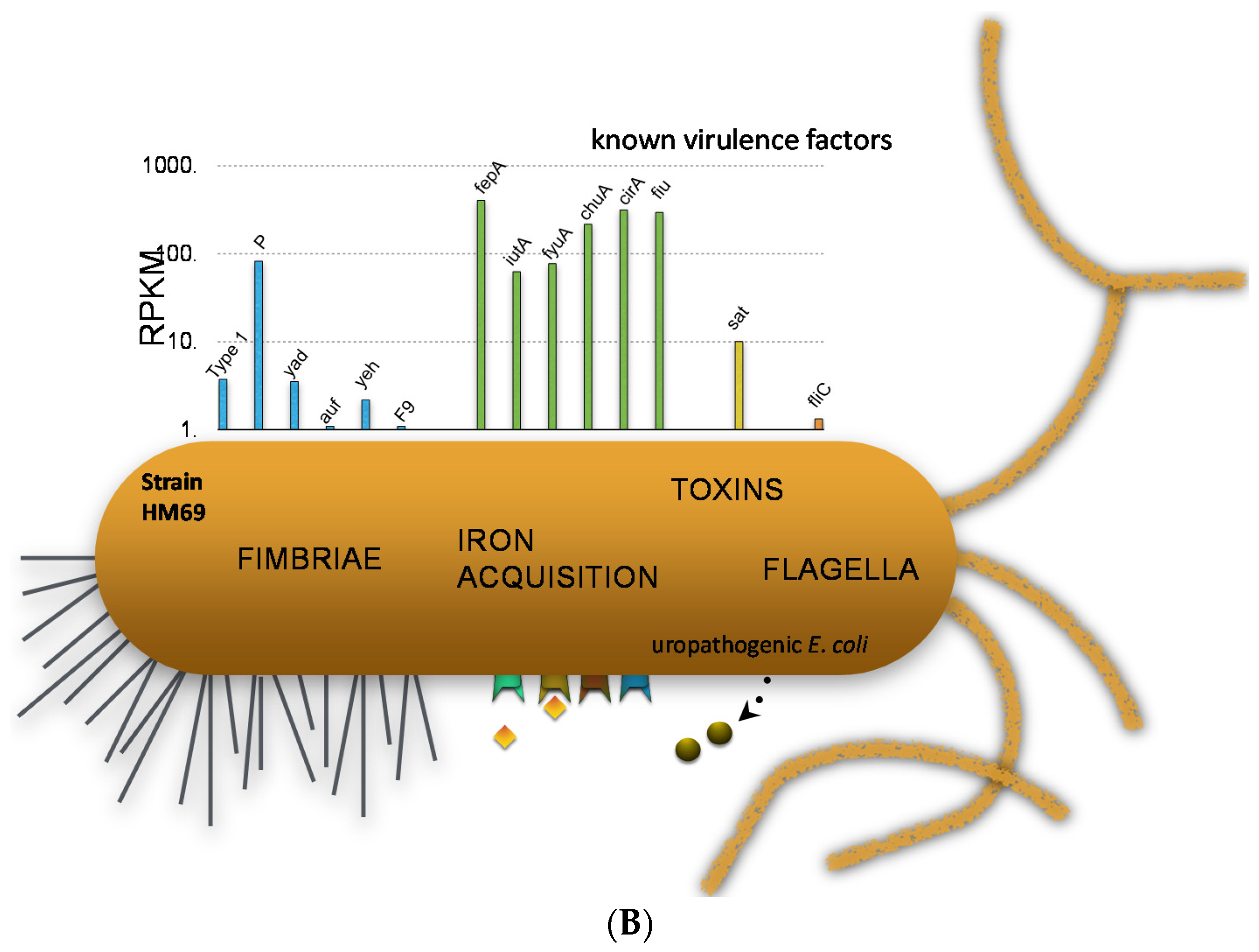
7. Comparative Transcriptomics Reveal Host-Induced Bacterial Genes
8. Different Bacterial Genes are Required for Different Host Settings
9. Redefining Bacterial Virulence
Acknowledgments
Conflicts of Interest
References
- Hensel, M.; Shea, J.E.; Gleeson, C.; Jones, M.D.; Dalton, E.; Holden, D.W. Simultaneous identification of bacterial virulence genes by negative selection. Science 1995, 269, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Slauch, J.M.; Mahan, M.J.; Mekalanos, J.J. In vivo expression technology for selection of bacterial genes specifically induced in host tissues. Methods Enzymol. 1994, 235, 481–492. [Google Scholar] [PubMed]
- Handfield, M.; Brady, L.J.; Progulske-Fox, A.; Hillman, J.D. Iviat: A novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 2000, 8, 336–339. [Google Scholar] [CrossRef]
- Shimoyama, T.; Everett, S.M.; Dixon, M.F.; Axon, A.T.; Crabtree, J.E. Chemokine mrna expression in gastric mucosa is associated with helicobacter pylori caga positivity and severity of gastritis. J. Clin. Pathol. 1998, 51, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.; Yang, C.; Oshima, T.; Mori, H.; Shimizu, K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 2004, 70, 2354–2366. [Google Scholar] [CrossRef] [PubMed]
- Salmon, K.; Hung, S.P.; Mekjian, K.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli k12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003, 278, 29837–29855. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Badarinarayana, V.; Selinger, D.W.; Janse, D.; Church, G.M. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003, 13, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Camejo, A.; Buchrieser, C.; Couve, E.; Carvalho, F.; Reis, O.; Ferreira, P.; Sousa, S.; Cossart, P.; Cabanes, D. In vivo transcriptional profiling of listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009, 5, e1000449. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.R.; Virtaneva, K.; Porcella, S.F.; Gardner, D.J.; Long, R.D.; Welty, D.M.; Barry, W.T.; Johnson, C.A.; Parkins, L.D.; Wright, F.A.; et al. Analysis of the transcriptome of group a streptococcus in mouse soft tissue infection. Am. J. Pathol. 2006, 169, 927–942. [Google Scholar] [CrossRef] [PubMed]
- Haugen, B.J.; Pellett, S.; Redford, P.; Hamilton, H.L.; Roesch, P.L.; Welch, R.A. In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain cft073 dsda. Infect. Immun. 2007, 75, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Revel, A.T.; Talaat, A.M.; Norgard, M.V. DNA microarray analysis of differential gene expression in borrelia burgdorferi, the lyme disease spirochete. Proc. Natl. Acad. Sci. USA 2002, 99, 1562–1567. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.A.; Haugen, B.J.; Buckles, E.L.; Lockatell, C.V.; Johnson, D.E.; Donnenberg, M.S.; Welch, R.A.; Mobley, H.L. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 2004, 72, 6373–6381. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Marlow, D.; Palyada, K.; Naikare, H.; Panciera, R.; Whitworth, L.; Clarke, C. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of campylobacter jejuni. Infect. Immun. 2005, 73, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Talaat, A.M.; Lyons, R.; Howard, S.T.; Johnston, S.A. The temporal expression profile of mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 2004, 101, 4602–4607. [Google Scholar] [CrossRef] [PubMed]
- Talaat, A.M.; Ward, S.K.; Wu, C.W.; Rondon, E.; Tavano, C.; Bannantine, J.P.; Lyons, R.; Johnston, S.A. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: Insights from genome-wide transcriptional profiling. J. Bacteriol. 2007, 189, 4265–4274. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.L.; Torrero, M.; Wheeler, P.R.; Truman, R.W.; Yoder, M.; Morrison, N.; Bishai, W.R.; Gillis, T.P. Biological implications of mycobacterium leprae gene expression during infection. J. Mol. Microbiol. Biotechnol. 2004, 8, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.R.; Marcus, E.A.; Wen, Y.; Oh, J.; Sachs, G. Gene expression in vivo shows that helicobacter pylori colonizes an acidic niche on the gastric surface. Proc. Natl. Acad. Sci. USA 2007, 104, 7235–7240. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.T.; Dolganov, N.A.; Otto, G.; Miller, M.C.; Wu, C.Y.; Schoolnik, G.K. Rpos controls the vibrio cholerae mucosal escape response. PLoS Pathog. 2006, 2, e109. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Dziejman, M.; Mekalanos, J.J. Determination of the transcriptome of vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Tuanyok, A.; Tom, M.; Dunbar, J.; Woods, D.E. Genome-wide expression analysis of burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect. Immun. 2006, 74, 5465–5476. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.L.; Puttamreddy, S.; Thacker, E.L.; Carruthers, M.D.; Minion, F.C. Transcriptome changes in mycoplasma hyopneumoniae during infection. Infect. Immun. 2008, 76, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Hagan, E.C.; Lloyd, A.L.; Rasko, D.A.; Faerber, G.J.; Mobley, H.L. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 2010, 6, e1001187. [Google Scholar] [CrossRef] [PubMed]
- Subashchandrabose, S.; Hazen, T.H.; Brumbaugh, A.R.; Himpsl, S.D.; Smith, S.N.; Ernst, R.D.; Rasko, D.A.; Mobley, H.L. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc. Natl. Acad. Sci. USA 2014, 111, 18327–18332. [Google Scholar] [CrossRef] [PubMed]
- Larocque, R.C.; Harris, J.B.; Dziejman, M.; Li, X.; Khan, A.I.; Faruque, A.S.; Faruque, S.M.; Nair, G.B.; Ryan, E.T.; Qadri, F.; et al. Transcriptional profiling of vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 2005, 73, 4488–4493. [Google Scholar] [CrossRef] [PubMed]
- Son, M.S.; Matthews, W.J., Jr.; Kang, Y.; Nguyen, D.T.; Hoang, T.T. In vivo evidence of pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 2007, 75, 5313–5324. [Google Scholar] [CrossRef] [PubMed]
- Timm, J.; Post, F.A.; Bekker, L.G.; Walther, G.B.; Wainwright, H.C.; Manganelli, R.; Chan, W.T.; Tsenova, L.; Gold, B.; Smith, I.; et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA 2003, 100, 14321–14326. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B.; Barlow, R.; D’Arcy, H.; Gillespie, B.; Sobel, J.D. Urinary tract infection: Self-reported incidence and associated costs. Ann. Epidemiol. 2000, 10, 509–515. [Google Scholar] [CrossRef]
- Lane, M.C.; Alteri, C.J.; Smith, S.N.; Mobley, H.L. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. USA 2007, 104, 16669–16674. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.A.; Burland, V.; Plunkett, G., 3rd; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.L.; Henderson, T.A.; Vigil, P.D.; Mobley, H.L. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 2009, 191, 3469–3481. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.L.; Rasko, D.A.; Mobley, H.L. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 2007, 189, 3532–3546. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Stapleton, A.E.; Johnson, J.R.; Walk, S.T.; Hooton, T.M.; Mobley, H.L. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: Contribution of ygi and yad fimbriae. Infect. Immun. 2011, 79, 4753–4763. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Himpsl, S.D.; Mobley, H.L. Preferential use of central metabolism in vivo reveals a nutritional basis for polymicrobial infection. PLoS Pathog. 2015, 11, e1004601. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Smith, S.N.; Mobley, H.L. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the tca cycle. PLoS Pathog. 2009, 5, e1000448. [Google Scholar] [CrossRef] [PubMed]
- Subashchandrabose, S.; Smith, S.N.; Spurbeck, R.R.; Kole, M.M.; Mobley, H.L. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 2013, 9, e1003788. [Google Scholar] [CrossRef] [PubMed]
- Falkow, S. Molecular koch’s postulates applied to microbial pathogenicity. Rev. Infect. Dis. 1988, 10, S274–S276. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mobley, H.L.T. Measuring Escherichia coli Gene Expression during Human Urinary Tract Infections. Pathogens 2016, 5, 7. https://doi.org/10.3390/pathogens5010007
Mobley HLT. Measuring Escherichia coli Gene Expression during Human Urinary Tract Infections. Pathogens. 2016; 5(1):7. https://doi.org/10.3390/pathogens5010007
Chicago/Turabian StyleMobley, Harry L. T. 2016. "Measuring Escherichia coli Gene Expression during Human Urinary Tract Infections" Pathogens 5, no. 1: 7. https://doi.org/10.3390/pathogens5010007




