Developments in Point-of-Care Diagnostic Technology for Cancer Detection
Abstract
:1. Introduction: Point-of-Care Diagnostic Tools
2. Cancer and the Glycome
3. POC Diagnostics for Cancer Detection
3.1. Lateral Flow Immunoassays
3.2. Circulating Tumor Cells
3.3. Prostate Cancer
3.4. Pancreatic Cancer
4. Emerging Biomarkers of Cancer and Their Use in POC Diagnostics
4.1. Autoantibody-Based Diagnostics
4.2. Exosomes
4.3. Lectin-Based Diagnostics
5. Future Directions
6. Discussion
Funding
Acknowledgments
Conflicts of Interest
References
- Song, Y.; Huang, Y.Y.; Liu, X.; Zhang, X.; Ferrari, M.; Qin, L. Point-of-care technologies for molecular diagnostics using a drop of blood. Trends Biotechnol. 2014, 32, 132–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uludag, Y.; Narter, F.; Sağlam, E.; Köktürk, G.; Gök, M.Y.; Akgün, M.; Barut, S.; Budak, S. An integrated lab-on-a-chip-based electrochemical biosensor for rapid and sensitive detection of cancer biomarkers. Anal. Bioanal. Chem. 2016, 408, 7775–7783. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Ng, S.R.; Xu, Y.; Dong, H.; Wang, Y.J.; Li, C.M. Advances of lab-on-a-chip in isolation, detection and post-processing of circulating tumour cells. Lab Chip 2013, 13, 3163–3182. [Google Scholar] [CrossRef] [PubMed]
- Zapatero Rodriguez, J.; O’Kennedy, R. New approaches for the develoment of diagnostic systems for prostate cancer. Asian Hosp. Healthc. Manag. 2017, 18–23. [Google Scholar]
- Pai, M.; Ghiasi, M.; Pant Pai, N. Point-of-care diagnostic testing in global health: What is the point? Microbe 2015, 10, 103–107. [Google Scholar] [CrossRef]
- Conroy, P.J.; Hearty, S.; Leonard, P.; O’Kennedy, R.J. Antibody production, design and use for biosensor-based applications. Semin. Cell Dev. Biol. 2009, 20, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Cho, R.J.; Gray, J.W. What are we learning from the cancer genome? Nat. Rev. Clin Oncol. 2012, 9, 621–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, K.; Tatsuguchi, A.; Saichi, N.; Toyama, A.; Tamura, K.; Furihata, M.; Takata, R.; Akamatsu, S.; Igarashi, M.; Nakayama, M.; et al. Plasma low-molecular-weight proteome profiling identified neuropeptide-Y as a prostate cancer biomarker polypeptide. J. Proteome Res. 2013, 12, 4497–4506. [Google Scholar] [CrossRef] [PubMed]
- Biskup, K.; Braicu, E.I.; Sehouli, J.; Fotopoulou, C.; Tauber, R.; Berger, M.; Blanchard, V. Serum glycome profiling: A biomarker for diagnosis of ovarian cancer. J. Proteome Res. 2013, 12, 4056–4063. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ruhaak, L.R.; Nguyen, U.T.; Taylor, S.L.; Dimapasoc, L.; Williams, C.; Stroble, C.; Ozcan, S.; Miyamoto, S.; Lebrilla, C.B.; et al. Evaluation of glycomic profiling as a diagnostic biomarker for epithelial ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Yu, S.-L.; Li, K.-C.; Yang, P.-C. Biomarkers and transcriptome profiling of lung cancer. Respirology 2012, 17, 620–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, V.J.; Weiss, R.H. Metabolomic profiling for early cancer detection: Current status and future prospects. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef] [PubMed]
- BEST (Biomarkers, EndpointS, and other Tools) Resource; FDA-NIH Biomarker Working Group Food and Drug Administration (US): Silver Spring, MD, USA, 2016.
- Cheah, A.L.; Goldblum, J.R.; Billings, S.D. Molecular diagnostics complementing morphology in superficial mesenchymal tumors. Semin. Diagn. Pathol. 2013, 30, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.J.; Wan, X.B.; Fu, X.H.; Wu, P.H.; Chen, D.K.; Wang, P.N.; Jiang, L.; Wang, D.H.; Chen, Z.T.; Huang, Y.; et al. Phosphorylated p38, a negative prognostic biomarker, complements TNM staging prognostication in colorectal cancer. Tumour Biol. 2014, 35, 10487–10495. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.R.; Ribeiro-Silva, A. Prognostic significance of immunohistochemical biomarkers in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2011, 40, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M.; Pitteri, S.J.; Faca, V.M. Mining the plasma proteome for cancer biomarkers. Nature 2008, 452, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Foj, L. Emerging biomarkers in the detection and prognosis of prostate cancer. Clin. Chem. Lab. Med. 2015, 53, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef] [PubMed]
- Lawler, K.; Kijanka, G. Clinical applications of immunoassays. In Immunoassays, Development, Applications and Future Trends, 1 ed.; O’Kennedy, R., Murphy, C., Eds.; Pan Stanford Publishing: New York, NY, USA, 2017; Volume 1, pp. 161–190. [Google Scholar]
- Plasma and Serum Preparation Protocols. Available online: https://www.thermofisher.com/in/en/home/references/protocols/cell-and-tissue-analysis/elisa-protocol/elisa-sample-preparation-protocols/plasma-and-serum-preparation.html (accessed on 18 May 2018).
- Salazar, C.; Nagadia, R.; Pandit, P.; Cooper-White, J.; Banerjee, N.; Dimitrova, N.; Coman, W.B.; Punyadeera, C. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell. Oncol. 2014, 37, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef] [PubMed]
- Bradwell, A.R.; Carr-Smith, H.D.; Mead, G.P.; Harvey, T.C.; Drayson, M.T. Serum test for assessment of patients with Bence Jones myeloma. Lancet 2003, 361, 489–491. [Google Scholar] [CrossRef]
- Liang, L.G.; Kong, M.Q.; Zhou, S.; Sheng, Y.F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.J.; Demirci, U.; et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arcaro, K.F.; Browne, E.P.; Qin, W.; Zhang, K.; Anderton, D.L.; Sauter, E.R. Differential expression of cancer-related proteins in paired breast milk samples from women with breast cancer. J. Hum. Lact. 2012, 28, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Melisko, M.E.; Magbanua, M.J.; Kablanian, A.T.; Scott, J.H.; Rugo, H.S.; Park, J.W. Detection of cerebrospinal fluid tumor cells and its clinical relevance in leptomeningeal metastasis of breast cancer. Breast Cancer Res. Treat. 2015, 154, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.J.; Murray, A.; McElveen, J.E.; Sahin, U.; Luxemburger, U.; Türeci, O. Autoantibodies in lung cancer: Possibilities for early detection and subsequent cure. Thorax 2008, 63, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, P.; Li, Z.; Xu, W.; Dai, L.; Wang, K.; Zhang, J. Evaluation of tumour-associated antigen (TAA) miniarray in immunodiagnosis of colon cancer. Scand. J. Immunol. 2009, 69, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zaenker, P.; Ziman, M.R. Serologic autoantibodies as diagnostic cancer biomarkers—A review. Cancer Epidemiol. Biomarkers Prev. 2013, 22, 2161–2181. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, J.-A.; Fitzgerald, J.; Fitzgerald, S.; Kenny, D.; Kay, E.W.; O’Kennedy, R.; Kijanka, G.S. Diagnostic potential of zinc finger protein-specific autoantibodies and associated linear B-cell epitopes in colorectal cancer. PLoS ONE 2015, 10, e0123469. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Kim, S.C.; Kim, H.J.; Ju, W.; Kim, Y.H.; Kim, H.J. A lectin-based diagnostic system using circulating antibodies to detect cervical intraepithelial neoplasia and cervical cancer. Glycobiology 2016, 26, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Tajiri, M.; Ohyama, C.; Wada, Y. Oligosaccharide profiles of the prostate specific antigen in free and complexed forms from the prostate cancer patient serum and in seminal plasma: A glycopeptide approach. Glycobiology 2008, 18, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Crawley, A.S.; O’Kennedy, R.J. The need for effective pancreatic cancer detection and management: A biomarker-based strategy. Expert Rev. Mol. Diagn. 2015, 15, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, L.; Sonpavde, G. Circulating biomarkers in bladder cancer. Bladder Cancer 2016, 4, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Moon, A. Identification of biomarkers for breast cancer using databases. J. Cancer Prev. 2016, 4, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Holdenrieder, S.; Pagliaro, L.; Morgenstern, D.; Dayyani, F. Clinically meaningful use of blood tumour markers in oncology. Biomed. Red. Int. 2016, 2016, 9795269. [Google Scholar]
- Elsafi, S.H.; Alqahtani, N.I.; Zakary, N.Y.; Al Zahrani, E.M. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clin. Exp. Gastroenterol. 2015, 8, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Bondgaard, A.L.; Høgdall, E.; Mellemgaard, A.; Skov, B.G. High specificity but low sensitivity of mutation-specific antibodies against EGFR mutations in non-small-cell lung cancer. Mod. Pathol. 2014, 27, 1590–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apweiler, R.; Hermjakob, H.; Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the swiss-prot database. Biochim. Biophys. Acta 1999, 1473, 4–8. [Google Scholar] [CrossRef]
- Fernandes, B.; Sagman, U.; Auger, M.; Demetrio, M.; Dennis, J.W. Beta 1-6 branched oligosaccharides as a marker of tumor progression in human breast and colon neoplasia. Cancer Res. 1991, 51, 718–723. [Google Scholar] [PubMed]
- Handerson, T.; Camp, R.; Harigopal, M.; Rimm, D.; Pawelek, J. Β1,6-branched oligosaccharides are increased in lymph node metastases and predict poor outcome in breast carcinoma. Clin. Cancer Res. 2005, 11, 2969–2973. [Google Scholar] [CrossRef] [PubMed]
- Burchell, J.; Poulsom, R.; Hanby, A.; Whitehouse, C.; Cooper, L.; Clausen, H.; Miles, D.; Taylor-Papadimitriou, J. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology 1999, 9, 1307–1311. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xie, X.; Liu, Y.; He, J.; Benitez, R.; Buckanovich, R.J.; Lubman, D.M. Identification and confirmation of differentially expressed fucosylated glycoproteins in the serum of ovarian cancer patients using a lectin array and LC-MS/MS. J. Proteome Res. 2012, 11, 4541–4552. [Google Scholar] [CrossRef] [PubMed]
- Peracaula, R.; Tabares, G.; Royle, L.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M.; de Llorens, R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology 2003, 13, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Misonou, Y.; Shida, K.; Korekane, H.; Seki, Y.; Noura, S.; Ohue, M.; Miyamoto, Y. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: Elucidation of aberrant glycosylation and its mechanistic causes in colorectal cancer cells. J. Proteome Res. 2009, 8, 2990–3005. [Google Scholar] [CrossRef] [PubMed]
- PSA Semi-Quantitative Rapid Test. Available online: http://ctkbiotech.com/ctk-product/psa-semi-quantitative-rapid-test/ (accessed on 21 May 2018).
- OncoE6 Cervical Test. Available online: http://www.arborvita.com/oncoe6/ (accessed on 22 May 2018).
- Chan, S.L.; Mo, F.; Johnson, P.J.; Siu, D.Y.W.; Chan, M.H.M.; Lau, W.Y.; Lai, P.B.S.; Lam, C.W.K.; Yeo, W.; Yu, S.C.H. Performance of serum α-fetoprotein levels in the diagnosis of hepatocellular carcinoma in patients with a hepatic mass. HPB 2014, 16, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zapatero-Rodríguez, J.; Estrela, P.; O’Kennedy, R. Point-of-care diagnostics in low resource settings: Present status and future role of microfluidics. Biosensors 2015, 5, 577–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.P. Circulating tumor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 4861. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Cellsearch. Available online: https://www.cellsearchctc.com/ (accessed on 21 March 2018).
- Riethdorf, S.; Fritsche, H.; Muller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Janicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the cellsearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Issadore, D.; Chung, J.; Shao, H.; Liong, M.; Ghazani, A.A.; Castro, C.M.; Weissleder, R.; Lee, H. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci. Transl. Med. 2012, 4, 141ra192. [Google Scholar] [CrossRef] [PubMed]
- Issadore, D. Point-of-care rare cell cancer diagnostics. Methods Mol. Biol. 2015, 1256, 123–137. [Google Scholar] [PubMed]
- Kirby, D.; Glynn, M.; Kijanka, G.; Ducree, J. Rapid and cost-efficient enumeration of rare cancer cells from whole blood by low-loss centrifugo-magnetophoretic purification under stopped-flow conditions. Cytometry Part A 2015, 87, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Mohamadi, R.M.; Poudineh, M.; Ahmed, S.U.; Ivanov, I.; Huang, C.L.; Moosavi, M.; Sargent, E.H.; Kelley, S.O. Single-cell mRNA cytometry via sequence-specific nanoparticle clustering and trapping. Nat. Chem. 2018, 10, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F.; Kumar, C.V.; Silvio Gutkind, J.; Patel, V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst 2010, 135, 2496–2511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; He, Y.; Wei, J.; Que, L. Nanostructured optical microchips for cancer biomarker detection. Biosens. Bioelectron. 2012, 38, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Irish Cancer Society: Screening for Prostate Cancer. Available online: https://www.cancer.ie/reduce-your-risk/health-education/cancer-awareness-campaigns/prostate-cancer-awareness/screening#sthash.KGk95gzs.dpbs (accessed on 20 February 2018).
- Hori, S.; Blanchet, J.S.; McLoughlin, J. From prostate-specific antigen (PSA) to precursor PSA (proPSA) isoforms: A review of the emerging role of proPSAs in the detection and management of early prostate cancer. BJU Int. 2013, 112, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Giménez, N. Evaluation of [−2] proPSA and prostate health index (phi) for the detection of prostate cancer: A systematic review and meta-analysis. Clin. Chem. Lab. Med. 2013, 51, 729. [Google Scholar] [CrossRef] [PubMed]
- Orava, E.W.; Cicmil, N.; Gariepy, J. Delivering cargoes into cancer cells using DNA aptamers targeting internalized surface portals. Biochim. Biophys. Acta 2010, 1798, 2190–2200. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.; Tharmalingam, T.; Rudd, P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Pihikova, D.; Kasak, P.; Tkac, J. Glycoprofiling of cancer biomarkers: Label-free electrochemical lectin-based biosensors. Open Chem. 2015, 13, 636–655. [Google Scholar] [CrossRef] [PubMed]
- Tabares, G.; Radcliffe, C.M.; Barrabes, S.; Ramirez, M.; Aleixandre, R.N.; Hoesel, W.; Dwek, R.A.; Rudd, P.M.; Peracaula, R.; de Llorens, R. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology 2006, 16, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Damborsky, P.; Madaboosi, N.; Soares, R.R.; Chu, V.; Conde, J.P.; Katrlik, J.; Estrela, P. DNA aptamer-based sandwich microfluidic assays for dual quantification and multi-glycan profiling of cancer biomarkers. Biosens. Bioelectron. 2016, 79, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, T.L.; McCubrey, J.A. Pancreatic cancer stem cells: Association with cell surface markers, prognosis, resistance, metastasis and treatment. Adv. Biol. Regul. 2014, 56, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Distler, M.; Aust, D.; Weitz, J.; Pilarsky, C.; Grutzmann, R. Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. BioMed Res. Int. 2014, 2014, 474905. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.W.; Gentry-Maharaj, A.; Fourkala, E.O.; Dawnay, A.; Burnell, M.; Zaikin, A.; Pedersen, A.E.; Jacobs, I.; Menon, U.; Wandall, H.H. Early detection of cancer in the general population: A blinded case-control study of p53 autoantibodies in colorectal cancer. Br. J. Cancer 2013, 108, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Karjalainen, A.; Koskinen, H.; Hemminki, K.; Vainio, H.; Shnaidman, M.; Ying, Z.; Pukkala, E.; Brandt-Rauf, P.W. P53 autoantibodies predict subsequent development of cancer. Int. J. Cancer 2005, 114, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.M.; Zhang, J. Autoantibodies to tumor-associated antigens: Reporters from the immune system. Immunol. Rev. 2008, 222, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M. Globocan v1.0, Cancer Incidence and Mortality Worldwide: Iarc Cancerbase No. 11; Intl Agency for Res on Cancer: Lyon, France, 2013; Volume 11. [Google Scholar]
- Burch, J.A.; Soares-Weiser, K.; St John, D.J.; Duffy, S.; Smith, S.; Kleijnen, J.; Westwood, M. Diagnostic accuracy of faecal occult blood tests used in screening for colorectal cancer: A systematic review. J. Med. Screen. 2007, 14, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Casiano, C.A.; Peng, X.X.; Koziol, J.A.; Chan, E.K.; Tan, E.M. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol. Biomarkers Prev. 2003, 12, 136–143. [Google Scholar] [PubMed]
- Kijanka, G.; Hector, S.; Kay, E.W.; Murray, F.; Cummins, R.; Murphy, D.; MacCraith, B.D.; Prehn, J.H.; Kenny, D. Human IgG antibody profiles differentiate between symptomatic patients with and without colorectal cancer. Gut 2010, 59, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, J.; Mange, A.; Solassol, J. Use of autoantibodies to detect the onset of breast cancer. J. Immunol. Res. 2014, 2014, 574981. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Su, G.L.; Wei, W.; Emick, D.; Conjeevaram, H.S.; Fontana, R.J.; Lok, A.S. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology 2003, 37, 1114–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, L.L.; Hua, P.Y.; Ye, L.L.; Wang, Y.C.; Qiu, T.; Bao, H.Z.; Wang, L. The detection of serum anti-p53 antibodies from patients with gastric carcinoma in china. Cancer Detect. Prev. 2007, 31, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W.S. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome mediated communication within the tumor microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through met. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Organ, S.L.; Tsao, M.-S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011, 3, S7–S19. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias Rde, O.; Machado Ldos, S.; Migliolo, L.; Franco, O.L. Insights into animal and plant lectins with antimicrobial activities. Molecules 2015, 20, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.A.; Carvalho, F.C.; Ruas, L.P.; Ricci-Azevedo, R.; Roque-Barreira, M.C. The immunomodulatory effect of plant lectins: A review with emphasis on ArtinM properties. Glycoconj. J. 2013, 30, 641–657. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kawagishi, H. Fungal lectins: A growing family. Methods Mol. Biol. 2014, 1200, 15–38. [Google Scholar] [PubMed]
- Ashwell, G.; Harford, J. Carbohydrate-specific receptors of the liver. Annu. Rev. Biochem. 1982, 51, 531–554. [Google Scholar] [CrossRef] [PubMed]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Dong, X.W.; Guo, X.L. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomed. Pharmacother. 2015, 69, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Monsigny, M.; Roche, A.C.; Kieda, C.; Midoux, P.; Obrenovitch, A. Characterization and biological implications of membrane lectins in tumor, lymphoid and myeloid cells. Biochimie 1988, 70, 1633–1649. [Google Scholar] [CrossRef]
- Ambrosi, M.; Cameron, N.R.; Davis, B.G. Lectins: Tools for the molecular understanding of the glycocode. Org. Biomol. Chem. 2005, 3, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Varrot, A.; Blanchard, B.; Imberty, A. Carbohydrate Recognition: Biological Problems, Methods and Applications; Wang, B., Boons, G.-J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Bray, F.; Loos, A.H.; McCarron, P.; Weiderpass, E.; Arbyn, M.; Moller, H.; Hakama, M.; Parkin, D.M. Trends in cervical squamous cell carcinoma incidence in 13 european countries: Changing risk and the effects of screening. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Lancucki, L.; Fender, M.; Koukari, A.; Lynge, E.; Mai, V.; Mancini, E.; Onysko, J.; Ronco, G.; Tornberg, S.; Vessey, M.; et al. A fall-off in cervical screening coverage of younger women in developed countries. J. Med. Screen. 2010, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, D.V.; Buck, C.B.; Pang, Y.Y.; Thompson, C.D.; Castle, P.E.; FitzGerald, P.C.; Kruger Kjaer, S.; Lowy, D.R.; Schiller, J.T. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 2004, 321, 205–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Franceschi, S.; Howell-Jones, R.; Snijders, P.J.; Clifford, G.M. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int. J. Cancer 2011, 128, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Zeng, Y.; Zeng, Y. Integrated microfluidic lectin barcode platform for high-performance focused glycomic profiling. Sci. Rep. 2016, 6, 20297. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.J.; Lo, J.H.; Bhatia, S.N. Smart nanosystems: Bio-inspired technologies that interact with the host environment. Proc. Natl. Acad. Sci. USA 2015, 112, 14460–14466. [Google Scholar] [CrossRef] [PubMed]
- Berguig, G.Y.; Convertine, A.J.; Frayo, S.; Kern, H.B.; Procko, E.; Roy, D.; Srinivasan, S.; Margineantu, D.H.; Booth, G.; Palanca-Wessels, M.C.; et al. Intracellular delivery system for antibody-peptide drug conjugates. Mol. Ther. 2015, 23, 907–917. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.E.; Yoon, J.-U. Recent approaches for optical smartphone sensing in resource-limited settings: A brief review. Anal. Methods 2016, 8, 6591–6601. [Google Scholar] [CrossRef]
- Wang, L.-J.; Chang, Y.-C.; Sun, R.; Li, L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens. Bioelectron. 2017, 87, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.; Stack, E.; Krivelo, S.; McPartlin, D.A.; Byrne, B.; Greef, C.; Lochhead, M.J.; Husar, G.; Devlin, S.; Elliott, C.T.; et al. Detection of the cyanobacterial toxin, microcystin-LR, using a novel recombinant antibody-based optical-planar waveguide platform. Biosens. Bioelectron. 2015, 67, 708–714. [Google Scholar] [CrossRef] [PubMed]

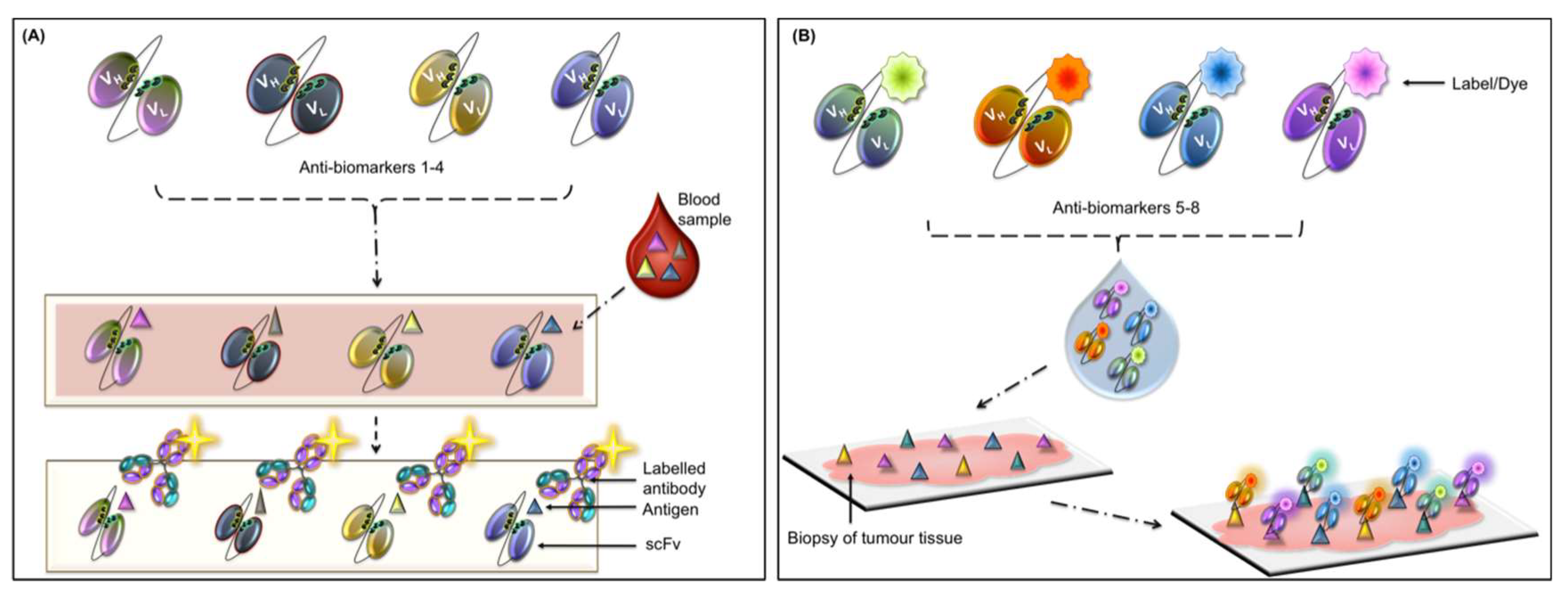
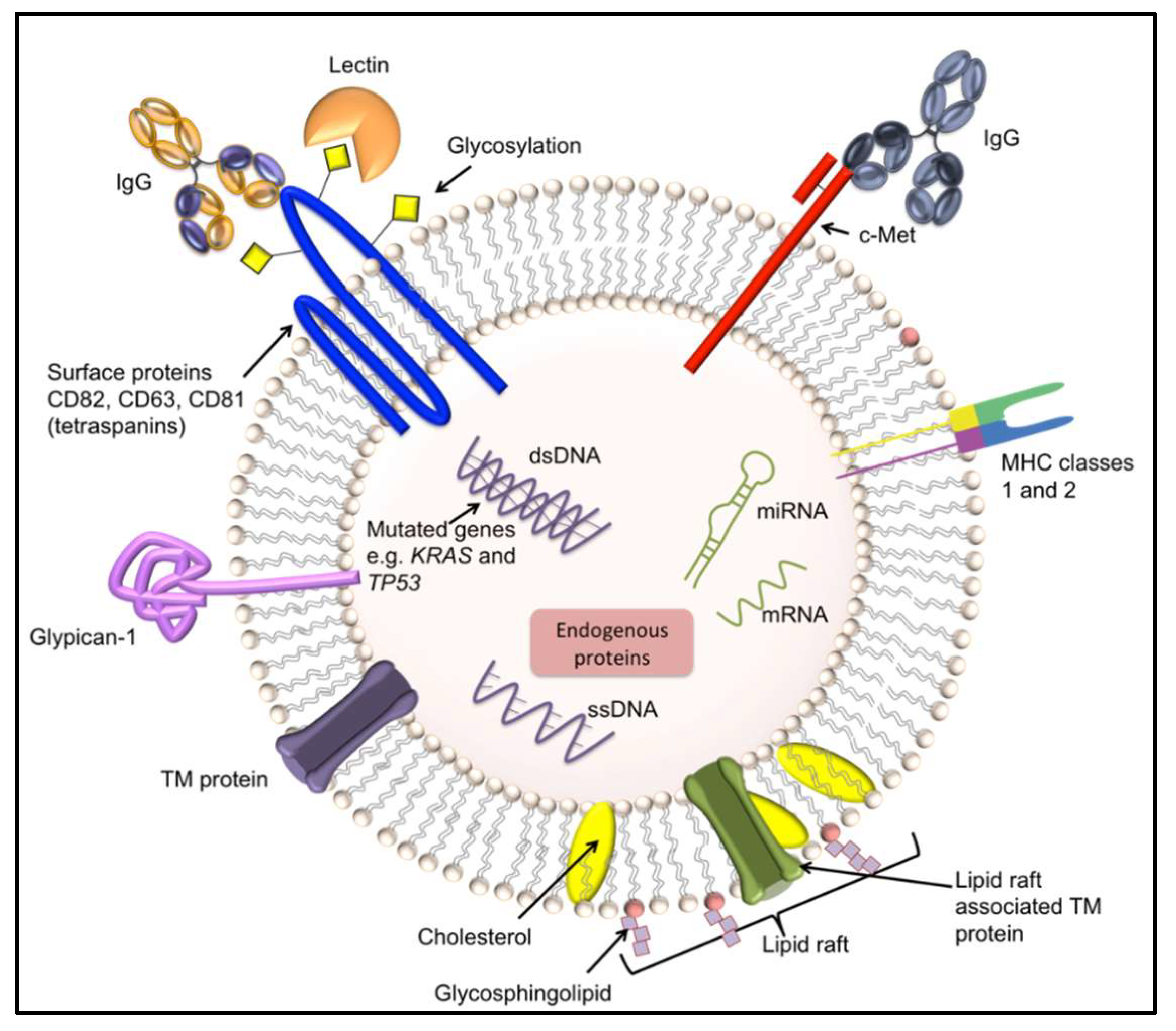
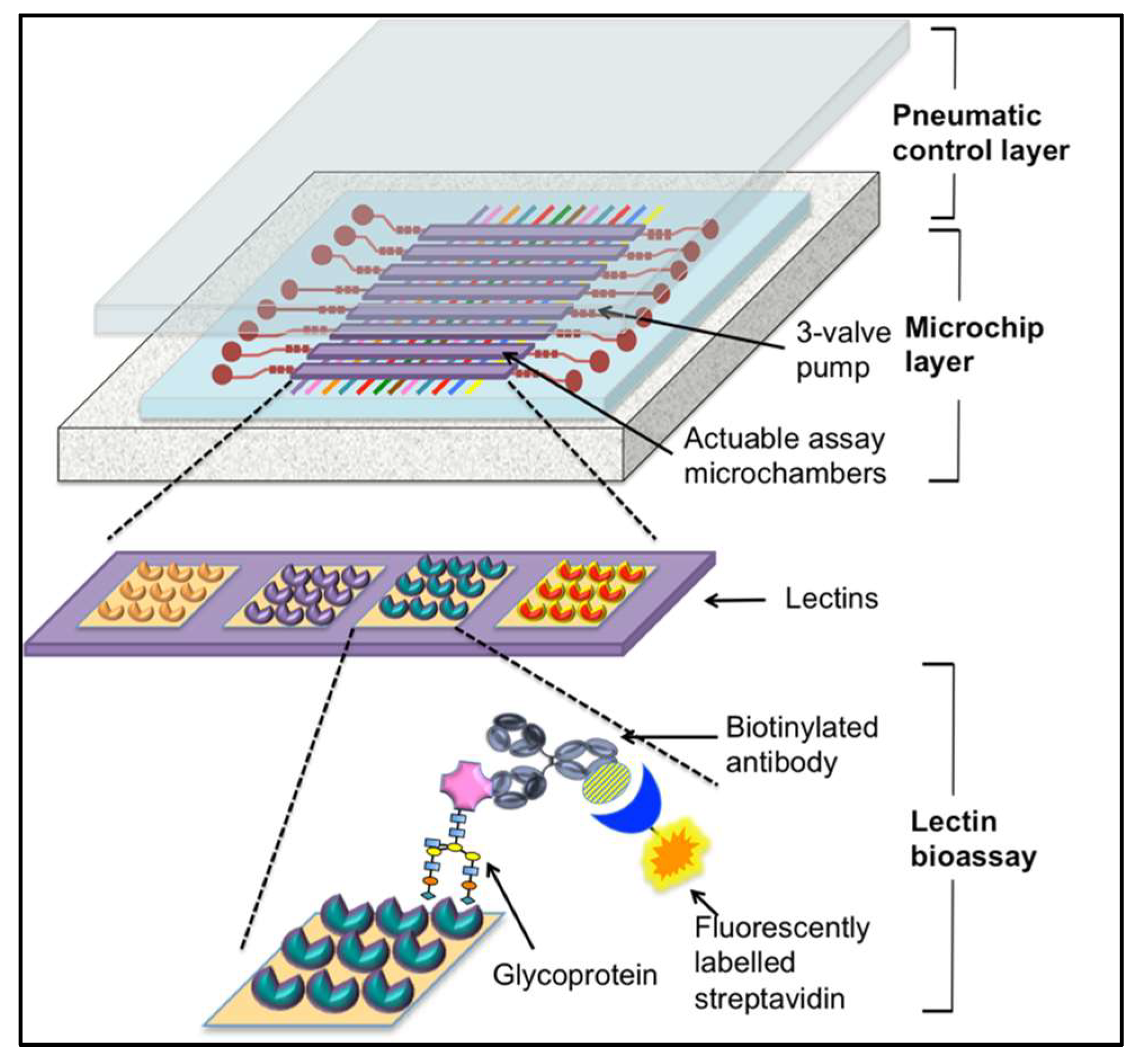
| Sample Matrix | Biomarkers | Associated Cancer | Ref. |
|---|---|---|---|
| Saliva | microRNA panel (miR-9, miR-134, miR-191) | Head and neck squamous cell carcinoma (HNSCC) | [24] |
| microRNA panel from whole saliva (miR-10b, miR-144, and miR-451), saliva supernatant (miR-10b, miR-144, miR-21, and miR-451) | Esophageal | [25] | |
| Urine | Bence Jones proteins | Light-chain multiple myeloma | [26] |
| Exosome size | Bladder | [27] | |
| Breast milk | TGF-β | Breast cancer | [28] |
| CSF | CTCs | Metastatic breast cancer giving rise to leptomeningeal metastasis | [29] |
| Serum from blood | PSA | Prostate | [2] |
| Autoantibodies | CRC, lung, stomach, breast | [30,31,32] | |
| ZNF | CRC | [33] | |
| Igs | CIN I and cervical cancer | [34] | |
| Seminal plasma | PSA | Prostate | [35] |
| Associated Cancer | Cancer Biomarker | POC Device | Clinical Capabilities | Test Duration | Sample | Company |
|---|---|---|---|---|---|---|
| Prostate | PSA | PSA Semi-quantitative rapid test | 4 ng/mL | 15 min | WB, S or P | CTK Biotech |
| Bladder | Nuclear matrix protein 22 (NMP 22) | Alere NMP22® BLADDERCHEK® | 99% Sensitivity when combined with cystoscopy | 30 min | Urine | Abbott (formerly Alere) |
| Colorectal | Fecal occult blood | FOB Rapid Test CE | hHB ≥ 50 ng/mL >98% specificity for hHB | 5–10 min | Stool | CTK Biotech |
| Cervical | OncoE6 | OncoE6™ Cervical Test | Sensitivity 84.6% Specificity 98.5% | 2.5 h | Cervical swab | Arbor Vita |
| HPV causing head and neck cancer | OncoE6 | OncoE6™ Oral Test | Still at testing stage | - | Oral swab | Arbor Vita |
| Liver | AFP | Medical IVD rapid diagnostic test kits AFP Test kit | Sensitivity 25 ng/mL Specificity 99% | 10 min | WB, S or P | INVBIO (Innovation Biotech) |
| Colorectal, breast, lung, | CEA | CEA Serum Rapid Test | 5 ng/mLSensitivity 97%, specificity 100% | 10 min | S or P | Cortez Diagnostics Inc. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, B.; Murphy, C.; Crawley, A.; O’Kennedy, R. Developments in Point-of-Care Diagnostic Technology for Cancer Detection. Diagnostics 2018, 8, 39. https://doi.org/10.3390/diagnostics8020039
Hayes B, Murphy C, Crawley A, O’Kennedy R. Developments in Point-of-Care Diagnostic Technology for Cancer Detection. Diagnostics. 2018; 8(2):39. https://doi.org/10.3390/diagnostics8020039
Chicago/Turabian StyleHayes, Bryony, Caroline Murphy, Aoife Crawley, and Richard O’Kennedy. 2018. "Developments in Point-of-Care Diagnostic Technology for Cancer Detection" Diagnostics 8, no. 2: 39. https://doi.org/10.3390/diagnostics8020039





