The Challenge to Stabilize, Extract and Analyze Urinary Cell-Free DNA (ucfDNA) during Clinical Routine
Abstract
:Highlights
- -
- Model system using urine spiked with synthetic cfDNA reference standard to compare various urine-stabilizing buffers under different storage conditions in analogy to the clinical setting.
- -
- Direct preservation with UAS showed the best results, ensuring sufficient ucfDNA quality for downstream analysis.
- -
- Specific detection of the EGFR variant T790M using ddPCR.
Abstract
1. Introduction
2. Material and Methods
2.1. Synthetic cfDNA Reference Standard with T790M
2.2. Collection and Preparation of Urine Samples from Healthy Donors
2.3. Choice of Urine Preservation Buffers for cfDNA Stabilization
2.4. Experimental Sample Setup to Mimic a Patient Sample during Clinical Routine
2.5. Isolation of cfDNA from Model Urine Samples
2.6. Fluorometric Quantification of ucfDNA
2.7. ucfDNA Quantification and Detection of T790M Variant Using ddPCR
2.8. Statistical Analysis
3. Results
3.1. Preliminary Tests
3.1.1. Volume of Reference Standard
3.1.2. Selection of Elution Volume
3.1.3. Concentration of Buffer AlloU
3.2. Analysis of ucfDNA in Healthy Urine Samples
3.3. Analysis of Model Urine Samples Spiked with cfDNA Reference Standard and Matching Negative Controls
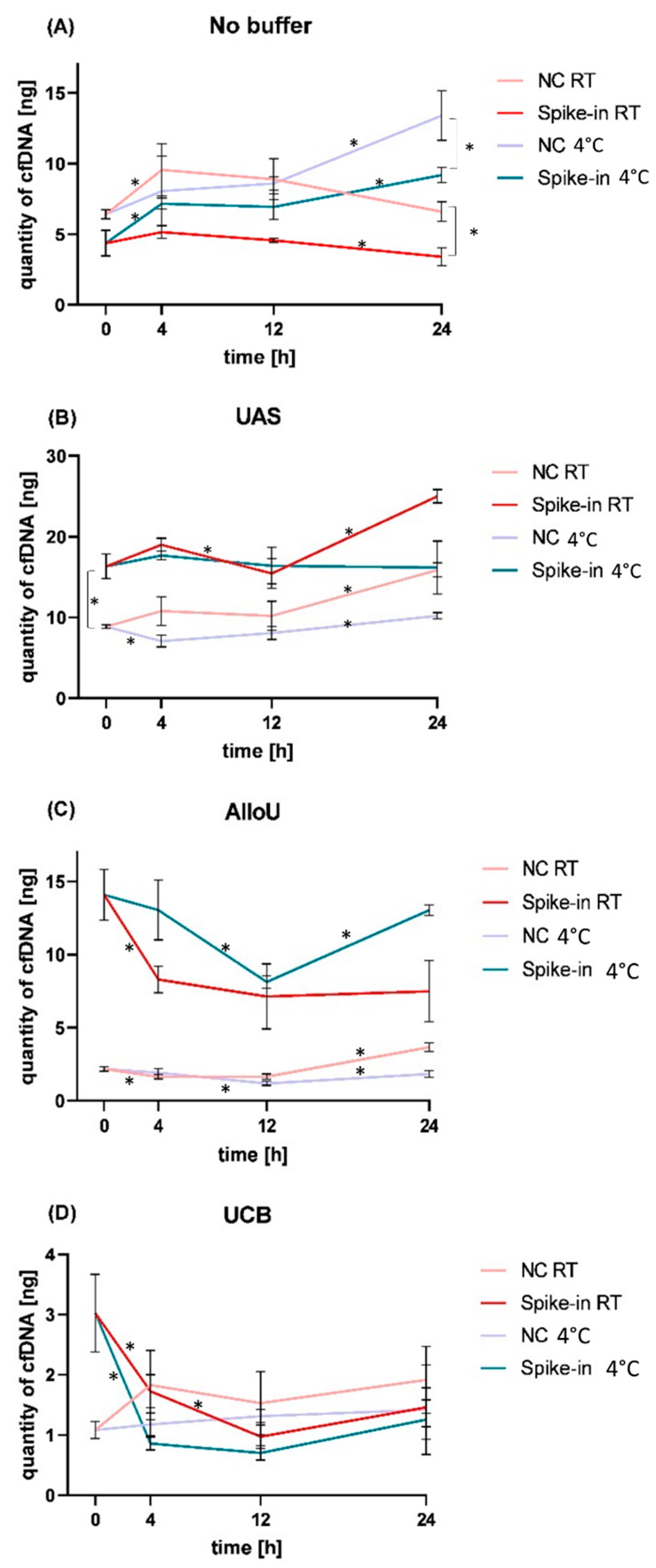
3.3.1. Storage without Preservation Buffer
3.3.2. Storage with UAS
3.3.3. Storage with AlloU
3.3.4. Storage with UCB
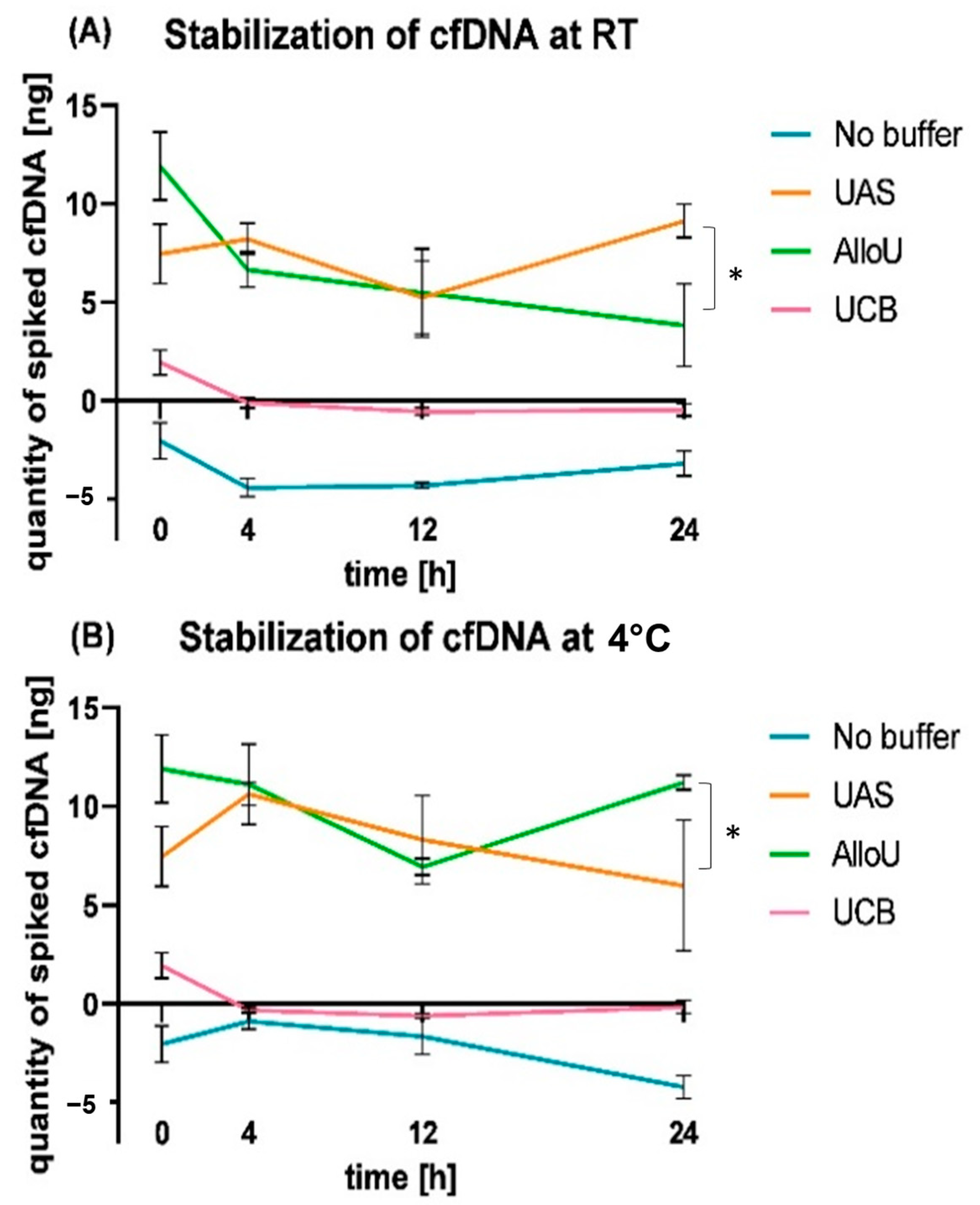
3.4. Relative cfDNA Dynamics in Spiked Urine Samples
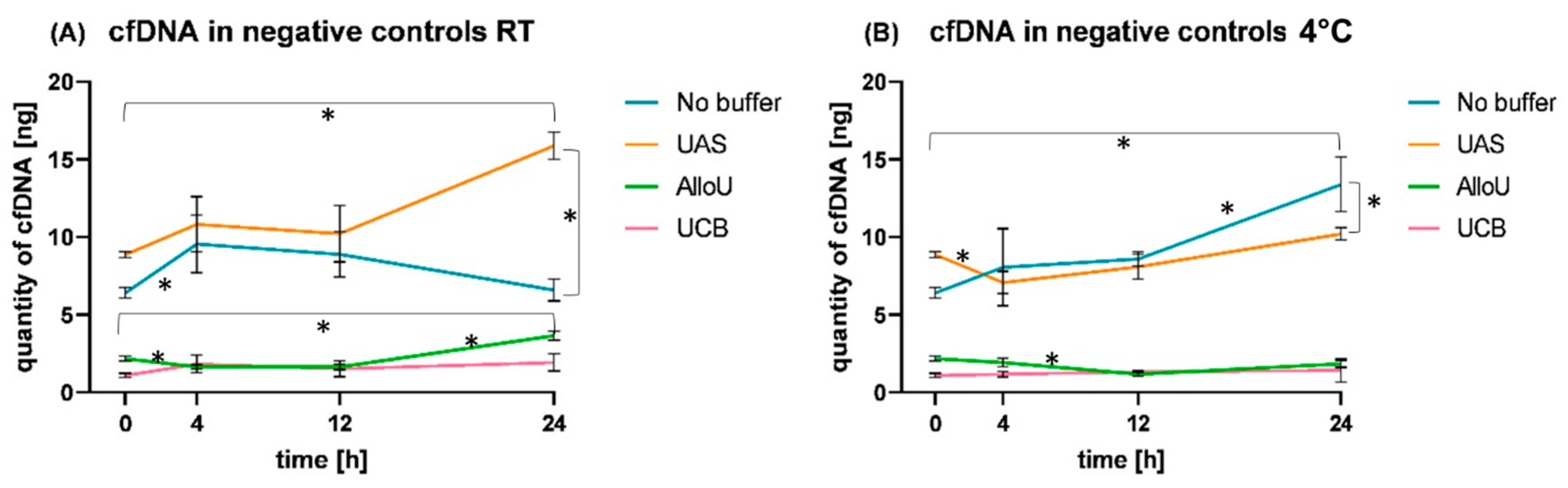
3.5. Observation of cfDNA Dynamics in NC Samples
3.6. Investigating the Concentration of Urinary cfDNA Using ddPCR vs. Qubit
3.7. Detecting the T790M Variant in Urinary cfDNA Using ddPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Contribution to the Field Statement
References
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Rolfo, C.; Cardona, A.F.; Cristofanilli, M.; Paz-Ares, L.; Mochon, J.J.; Duran, I.; Raez, L.E.; Russo, A.; Lorente, J.A.; Malapelle, U.; et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit. Rev. Oncol. Hematol. 2020, 151, 102978. [Google Scholar] [CrossRef]
- Delgado-Ureña, M.; Ortega, F.G.; de Miguel-Pérez, D.; Rodriguez-Martínez, A.; García-Puche, J.L.; Ilyine, H.; Lorente, J.A.; Exposito-Hernandez, J.; Garrido-Navas, M.C.; Delgado-Ramirez, M.; et al. Circulating tumor cells criteria (CyCAR) versus standard RECIST criteria for treatment response assessment in metastatic colorectal cancer patients. J. Transl. Med. 2018, 16, 251. [Google Scholar] [CrossRef]
- Botezatu, I.; Serdyuk, O.G.; Potapova, G.; Shelepov, V.; Alechina, R.; Molyaka, Y.; Anan’ev, V.; Bazin, I.; Garin, A.; Narimanov, M.; et al. Genetic Analysis of DNA Excreted in Urine: A New Approach for Detecting Specific Genomic DNA Sequences from Cells Dying in an Organism. Clin. Chem. 2000, 46, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, W. Association of urinary and plasma DNA in early breast cancer patients and its links to disease relapse. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2018, 20, 1053–1060. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, V.; Bronkhorst, A.J.; Van den Ackerveken, P.; Herzog, M.; Holdenrieder, S. Serial profiling of cell-free DNA and nucleosome histone modifications in cell cultures. Sci. Rep. 2021, 11, 9460. [Google Scholar] [CrossRef]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef]
- Salvi, S.; Casadio, V. Urinary Cell-Free DNA: Potential and Applications. Methods Mol. Biol. 2019, 1909, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Herzog, H.; Dogan, S.; Aktas, B.; Nel, I. Targeted Sequencing of Plasma-Derived vs. Urinary cfDNA from Patients with Triple-Negative Breast Cancer. Cancers 2022, 14, 4101. [Google Scholar] [CrossRef]
- Streleckiene, G.; Reid, H.M.; Arnold, N.; Bauerschlag, D.; Forster, M. Quantifying cell free DNA in urine: Comparison between commercial kits, impact of gender and inter-individual variation. BioTechniques 2018, 64, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.D.; Chan, K.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Su, Y.H.; Wang, M.; Brenner, D.E.; Ng, A.; Melkonyan, H.; Umansky, S.; Syngal, S.; Block, T.M. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J. Mol. Diagn. 2004, 6, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Laktionov, P.P. Extracellular Nucleic Acids in Urine: Sources, Structure, Diagnostic Potential. Acta Naturae 2015, 7, 48–54. [Google Scholar] [CrossRef]
- Hudecova, I.; Smith, C.G.; Hänsel-Hertsch, R.; Chilamakuri, C.S.; Morris, J.A.; Vijayaraghavan, A.; Heider, K.; Chandrananda, D.; Cooper, W.N.; Gale, D.; et al. Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res. 2022, 32, 215–227. [Google Scholar] [CrossRef]
- Lu, T.; Li, J. Clinical applications of urinary cell-free DNA in cancer: Current insights and promising future. Am. J. Cancer Res. 2017, 7, 2318–2332. [Google Scholar]
- Salvi, S.; Gurioli, G.; De Giorgi, U.; Conteduca, V.; Tedaldi, G.; Calistri, D.; Casadio, V. Cell-free DNA as a diagnostic marker for cancer: Current insights. OncoTargets Ther. 2016, 9, 6549–6559. [Google Scholar] [CrossRef]
- Millholland, J.M.; Li, S.; Fernandez, C.A.; Shuber, A.P. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res. Rep. Urol. 2012, 4, 33–40. [Google Scholar] [CrossRef]
- Nel, I.; Herzog, H.; Aktas, B. Combined Analysis of Disseminated Tumor Cells (DTCs) and Circulating Tumor DNA (ctDNA) in a Patient Suffering from Triple Negative Breast Cancer Revealed Elevated Risk. Front. Biosci. (Landmark Ed.) 2022, 27, 208. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Burness, M.L.; Grushko, T.A.; Olopade, O.I. Epidermal growth factor receptor in triple-negative and basal-like breast cancer: Promising clinical target or only a marker? Cancer J. 2010, 16, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Vaclova, T.; Grazini, U.; Ward, L.; O’Neill, D.; Markovets, A.; Huang, X.; Chmielecki, J.; Hartmaier, R.; Thress, K.S.; Smith, P.D.; et al. Clinical impact of subclonal EGFR T790M mutations in advanced-stage EGFR-mutant non-small-cell lung cancers. Nat. Commun. 2021, 12, 1780. [Google Scholar] [CrossRef]
- Ma, C.; Wei, S.; Song, Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J. Thorac. Dis. 2011, 3, 10–18. [Google Scholar] [CrossRef]
- Silveira, C.; Sousa, A.C.; Janeiro, A.; Malveiro, S.; Teixeira, E.; Brysch, E.; Pantarotto, M.; Felizardo, M.; Madureira, R.; Nogueira, F.; et al. Detection and quantification of EGFR T790M mutation in liquid biopsies by droplet digital PCR. Transl. Lung Cancer Res. 2021, 10, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Jordaens, S.; Mehta, A.; Faster, D.; Laeremans, M.; Bruyninckx, S.; Pasmans, D.; Doukhanine, E.; Beyers, K.; Vankerckhoven, V. White Paper—Why Urine Preservation Is Needed for Molecular Cancer Biomarker Detection. 2020. Available online: https://www.semanticscholar.org/paper/Why-urine-preservation-is-needed-for-molecular-Arora-Jordaens/e2e592daa7a71cf50f27da75312bb119da97c351 (accessed on 22 November 2023).
- Bosschieter, J.; Bach, S.; Bijnsdorp, I.V.; Segerink, L.I.; Rurup, W.F.; Van Splunter, A.P.; Bahce, I.; Novianti, P.W.; Kazemier, G.; Van Moorselaar, R.J.; et al. A protocol for urine collection and storage prior to DNA methylation analysis. PLoS ONE 2018, 13, e0200906. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, F.; Guo, L.; Chen, R.; Yuan, X.; Li, C.; Shu, L.; Liu, H.; Zhou, Y.; Wu, Y.; et al. A novel urine cell-free DNA preservation solution and its application in kidney transplantation. Nephrology 2021, 26, 684–691. [Google Scholar] [CrossRef]
- Barra, G.B.; Santa Rita, T.H.; de Almeida Vasques, J.; Chianca, C.F.; Nery, L.F.; Costa, S.S. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin. Biochem. 2015, 48, 976–981. [Google Scholar] [CrossRef]
- Yasuda, T.; Awazu, S.; Sato, W.; Iida, R.; Tanaka, Y.; Kishi, K. Human genetically polymorphic deoxyribonuclease: Purification, characterization, and multiplicity of urine deoxyribonuclease I. J. Biochem. 1990, 108, 393–398. [Google Scholar] [CrossRef]
- Moon, W.J.; Yang, Y.; Liu, J. Zn2+-Dependent DNAzymes: From Solution Chemistry to Analytical, Materials and Therapeutic Applications. Chembiochem Eur. J. Chem. Biol. 2021, 22, 779–789. [Google Scholar] [CrossRef]
- Amponin, D.E.; Przybek-Skrzypecka, J.; Zyablitskaya, M.; Takaoka, A.; Suh, L.H.; Nagasaki, T.; Trokel, S.L.; Paik, D.C. Ex vivo anti-microbial efficacy of various formaldehyde re-leasers against antibiotic resistant and antibiotic sensitive microorganisms involved in infectious keratitis. BMC Ophthalmol. 2020, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.V.; Hoeck, U.; Breinholdt, J.; Olsen, C.E.; Kreilgaard, B. Characterization and chemistry of imidazolidinyl urea and diazolidinyl urea. Contact Dermat. 2006, 54, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Bos, M.K.; Nasserinejad, K.; Jansen, M.P.; Steendam, C.M.; Angus, L.; Atmodimedjo, P.N.; de Jonge, E.; Dinjens, W.N.; van Schaik, R.H.; Del Re, M.; et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol. Oncol. 2021, 15, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Biorad Droplet Digital Applications Guide. Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (accessed on 7 February 2023).
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef]
- Augustus, E.; Van Casteren, K.; Sorber, L.; van Dam, P.; Roeyen, G.; Peeters, M.; Vorsters, A.; Wouters, A.; Raskin, J.; Rolfo, C.; et al. The art of obtaining a high yield of cell-free DNA from urine. PLoS ONE 2020, 15, e0231058. [Google Scholar] [CrossRef]
- Multiplex I cfDNA Reference Standard Set in Synthetic Plasma. 2022. Available online: https://horizondiscovery.com/en/reference-standards/products/multiplex-i-cfdna-reference-standard-set-in-synthetic-plasma#supporting-data (accessed on 22 November 2023).
- Einstein, A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905, 322, 549–560. [Google Scholar] [CrossRef]
- Zymo Research Europe. Urine Conditioning Buffer_Protocoll. 2022. Available online: https://files.zymoresearch.com/protocols/_d3061-1-8_d3061-1-140_urine_collection.pdf (accessed on 22 November 2023).
- Martignano, F. Cell-Free DNA: An Overview of Sample Types and Isolation Procedures. Methods Mol. Biol. 2019, 1909, 13–27. [Google Scholar] [CrossRef]
- Szarvas, T.; Kovalszky, I.; Bedi, K.; Szendroi, A.; Majoros, A.; Riesz, P.; Füle, T.; László, V.; Kiss, A.; Romics, I. Deletion analysis of tumor and urinary DNA to detect bladder cancer: Urine supernatant versus urine sediment. Oncol. Rep. 2007, 18, 405–409. [Google Scholar] [CrossRef]
- Palacín-Aliana, I.; García-Romero, N.; Asensi-Puig, A.; Carrión-Navarro, J.; González-Rumayor, V.; Ayuso-Sacido, Á. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines 2021, 9, 906. [Google Scholar] [CrossRef] [PubMed]

| Stabilizing Reagent | Sample Type | Urine Volume | Buffer Volume | Reference cfDNA Spike-In Volume |
|---|---|---|---|---|
| No buffer | NC | 3 mL | - | - |
| No buffer | spike | 3 mL | - | 60 µL |
| U A S | NC | 3 mL | 1 mL | - |
| U A S | spike | 3 mL | 1 mL | 60 µL |
| AlloU | NC | 3 mL | 120 µL | - |
| AlloU | spike | 3 mL | 120 µL | 60 µL |
| UCB | NC | 3 mL | 210 µL | - |
| UCB | spike | 3 mL | 210 µL | 60 µL |
| Sample | Total cfDNA/3 mL (ng) | Mean | SD | Level of Significance |
|---|---|---|---|---|
| F1 | 1.85 | |||
| 1.85 | 2.06 | 0.30 | ||
| 2.49 | ||||
| F2 | 61.17 | |||
| 74.13 | 73.53 | 9.86 | ||
| 85.30 | ||||
| F3 | 1.29 | |||
| 1.01 | 1.24 | 0.17 | ||
| 1.42 | ||||
| F4 | 1.62 | |||
| 1.95 | 2.19 | 0.60 | ||
| 3.01 | ||||
| Female cfDNA (ng/3 mL) | 19.76 | 31.44 | ||
| M1 | 0 | |||
| 0 | 0 | 0 | ||
| 0 | ||||
| M2 | 8.82 | |||
| 16.93 | 14.13 | 3.76 | ||
| 16.63 | ||||
| M3 | 1.11 | |||
| 1.38 | 1.32 | 0.16 | ||
| 1.48 | ||||
| M4 | 5.59 | |||
| 5.83 | 6.03 | 0.46 | ||
| 6.66 | ||||
| Male cfDNA (ng/3 mL) | 5.37 | 5.85 | p= 0.15 | |
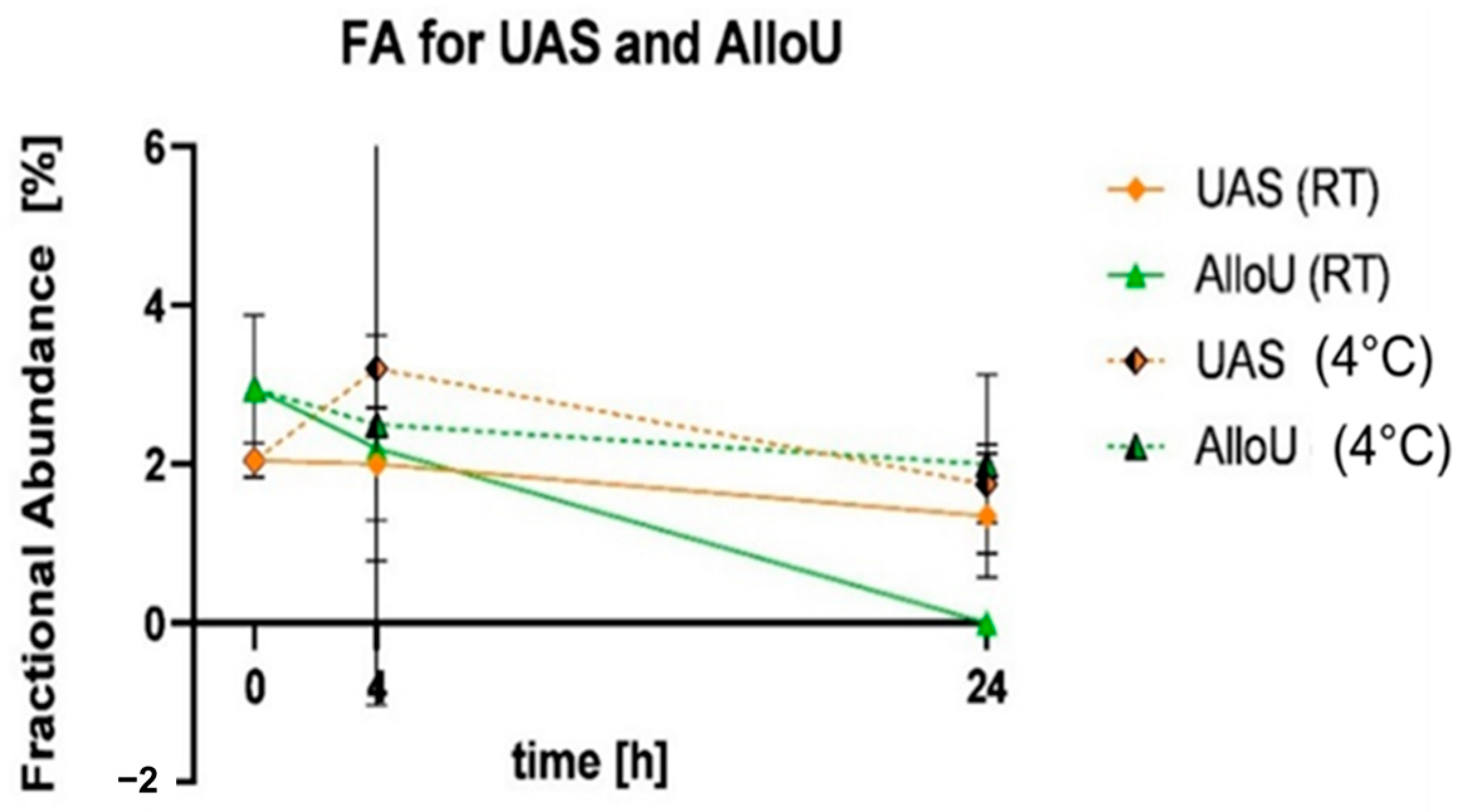
| Storage Conditions | Samples Buffered with UAS | Samples Buffered with AlloU | |||
|---|---|---|---|---|---|
| Time (h) | Temperature | Expected FA of Variant (%) | Measured FA of Variant (%) | Expected FA of Variant (%) | Measured FA of Variant (%) |
| 0 | 2.41 | 2.05 | 4.26 | 2.95 | |
| 4 | RT | 2.41 | 2.00 | 3.95 | 2.25 |
| 4 | 4 °C | 3.31 | 3.20 | 4.22 | 2.50 |
| 24 | RT | 1.96 | 1.90 | 2.65 | 0 |
| 24 | 4 °C | 1.25 | 1.80 | 4.39 | 2.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nel, I.; Münch, C.; Shamkeeva, S.; Heinemann, M.L.; Isermann, B.; Aktas, B. The Challenge to Stabilize, Extract and Analyze Urinary Cell-Free DNA (ucfDNA) during Clinical Routine. Diagnostics 2023, 13, 3670. https://doi.org/10.3390/diagnostics13243670
Nel I, Münch C, Shamkeeva S, Heinemann ML, Isermann B, Aktas B. The Challenge to Stabilize, Extract and Analyze Urinary Cell-Free DNA (ucfDNA) during Clinical Routine. Diagnostics. 2023; 13(24):3670. https://doi.org/10.3390/diagnostics13243670
Chicago/Turabian StyleNel, Ivonne, Carolin Münch, Saikal Shamkeeva, Mitja L. Heinemann, Berend Isermann, and Bahriye Aktas. 2023. "The Challenge to Stabilize, Extract and Analyze Urinary Cell-Free DNA (ucfDNA) during Clinical Routine" Diagnostics 13, no. 24: 3670. https://doi.org/10.3390/diagnostics13243670






