A Hybrid Technique for Diabetic Retinopathy Detection Based on Ensemble-Optimized CNN and Texture Features
Abstract
:1. Introduction
- We propose an efficient hybrid technique that uses an ensemble-optimized CNN for automated diabetic retinopathy detection to improve classification accuracy.
- We propose a novel GraphNet124 for feature extraction and to train a pretrained ResNet50 for diabetic retinopathy images, and then, the features are extracted using the transfer learning technique.
- We propose a feature fusion and selection approach that works in three steps: (i) features from GraphNet124 and ResNet50 are selected using Shannon Entropy, and then, fused; (ii) these fused features are optimized using the Binary Dragonfly Algorithm (BDA) [22] and the Sine Cosine Algorithm (SCA) [23]; (iii) the features extracted from LBP are also selected using Shannon Entropy, and then, fused with the optimized features found in step (ii).
- We evaluate the proposed hybrid architecture on a complex, publicly available, and standardized dataset (Kaggle EyePACS).
- We compare the performance of the proposed hybrid technique, including the fusion of discriminative features from GraphNet124, ResNet50, and LBP, with baseline techniques.
- To the best of our knowledge, this study is the first in the domain of DR abnormality detection and classification using the fusion of automated CNN-based features and LBP-based textural features.
2. Related Work
3. Proposed Methodology
3.1. Dataset
3.2. Preprocessing
3.3. Feature Engineering
3.3.1. LBP Feature Extraction
3.3.2. CNN Feature Extraction
3.3.3. Feature Selection and Fusion
4. Results and Discussion
4.1. Experimental Setup
4.2. Dataset
4.3. Performance Measures
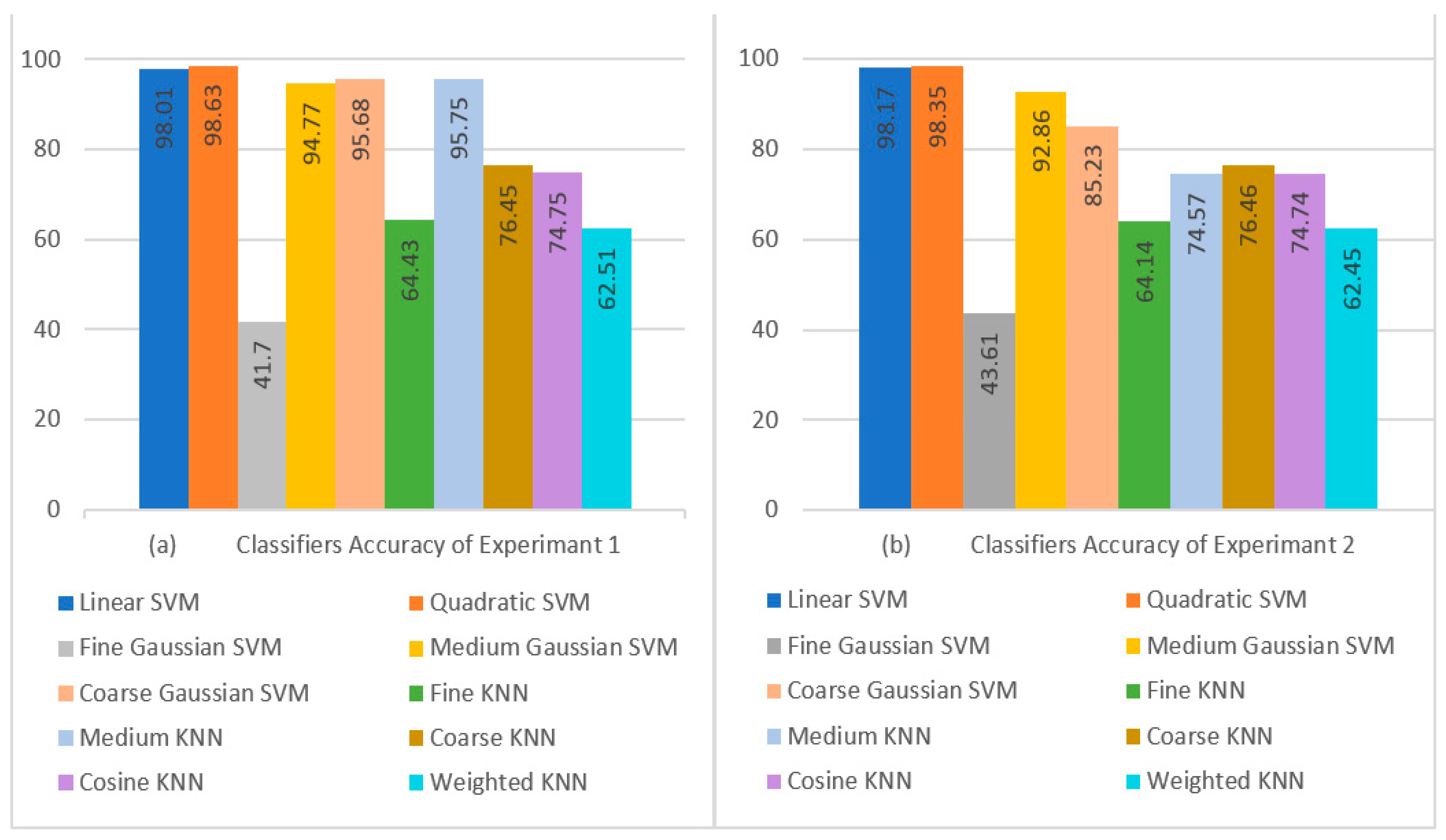
4.4. Experiment 1: Classification Results Using Feature Vector with Dimensions of and 5-Fold Cross-Validation
4.5. Experiment 2: Classification Results Using Feature Vector with Dimensions of and 5-Fold Cross-Validation
4.6. Experiment 3: Classification Results Using Feature Vector with Dimensions of and 10-Fold Cross-Validation
4.7. Experiment 4: Classification Results Using Feature Vector with Dimensions of and 10-Fold Cross-Validation
4.8. Comparison with Existing Methods
4.9. Quantitative Analysis of Proposed Method’s Average Performance
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- García, G.; Gallardo, J.; Mauricio, A.; López, J.; Carpio, C.D. Detection of diabetic retinopathy based on a convolutional neural network using retinal fundus images. In Proceedings of the International Conference on Artificial Neural Networks, Alghero, Italy, 11–14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Teo, Z.L.; Tham, Y.-C.; Yu, M.; Chee, M.L.; Rim, T.H.; Cheung, N.; Bikbov, M.M.; Wang, Y.X.; Tang, Y.; Lu, Y. Global prevalence of diabetic retinopathy and projection of burden through 2045: Systematic review and meta-analysis. Ophthalmology 2021, 128, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Kempen, J.H.; O’Colmain, B.J.; Leske, M.C.; Haffner, S.M.; Klein, R.; Moss, S.E.; Taylor, H.R.; Hamman, R.F. The prevalence of diabetic retinopathy among adults in the United States. Arch. Ophthalmol. (Chic. Ill.: 1960) 2004, 122, 552–563. [Google Scholar]
- Serrano, C.I.; Shah, V.; Abràmoff, M.D. Use of expectation disconfirmation theory to test patient satisfaction with asynchronous telemedicine for diabetic retinopathy detection. Int. J. Telemed. Appl. 2018, 2018, 7015272. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Dinh, A.V.; Wahid, K.A. Automated diabetic retinopathy detection using bag of words approach. J. Biomed. Sci. Eng. 2017, 10, 86–96. [Google Scholar] [CrossRef]
- Costa, P.; Galdran, A.; Smailagic, A.; Campilho, A. A weakly-supervised framework for interpretable diabetic retinopathy detection on retinal images. IEEE Access 2018, 6, 18747–18758. [Google Scholar] [CrossRef]
- Savastano, M.C.; Federici, M.; Falsini, B.; Caporossi, A.; Minnella, A.M. Detecting papillary neovascularization in proliferative diabetic retinopathy using optical coherence tomography angiography. Acta Ophthalmol. 2018, 96, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zhu, Y.; Zhou, H. Diabetic retinopathy detection using prognosis of microaneurysm and early diagnosis system for non-proliferative diabetic retinopathy based on deep learning algorithms. IEEE Access 2020, 8, 104292–104302. [Google Scholar] [CrossRef]
- Vashist, P.; Singh, S.; Gupta, N.; Saxena, R. Role of early screening for diabetic retinopathy in patients with diabetes mellitus: An overview. Indian J. Community Med. 2011, 36, 247–252. [Google Scholar] [CrossRef]
- Prentašić, P.; Lončarić, S. Detection of exudates in fundus photographs using deep neural networks and anatomical landmark detection fusion. Comput. Methods Programs Biomed. 2016, 137, 281–292. [Google Scholar] [CrossRef]
- Mahendran, G.; Dhanasekaran, R. Investigation of the severity level of diabetic retinopathy using supervised classifier algorithms. Comput. Electr. Eng. 2015, 45, 312–323. [Google Scholar] [CrossRef]
- Santhi, D.; Manimegalai, D.; Parvathi, S.; Karkuzhali, S. Segmentation and classification of bright lesions to diagnose diabetic retinopathy in retinal images. Biomed. Eng. Biomed. Tech. 2016, 61, 443–453. [Google Scholar] [CrossRef]
- Chudzik, P.; Majumdar, S.; Calivá, F.; Al-Diri, B.; Hunter, A. Microaneurysm detection using fully convolutional neural networks. Comput. Methods Programs Biomed. 2018, 158, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Yu, S.; Vignarajan, J.; An, D.; Tay-Kearney, M.-L.; Kanagasingam, Y. Retinal hemorrhage detection by rule-based and machine learning approach. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Republic of Korea, 11–15 July 2017. [Google Scholar]
- Jaya, T.; Dheeba, J.; Singh, N.A. Detection of Hard Exudates in Colour Fundus Images Using Fuzzy Support Vector Machine-Based Expert System. J. Digit. Imaging 2015, 28, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.; Palani, S. Hierarchical classifier for soft and hard exudates detection of retinal fundus images. J. Intell. Fuzzy Syst. 2014, 27, 2511–2528. [Google Scholar] [CrossRef]
- Zhou, W.; Wu, C.; Chen, D.; Yi, Y.; Du, W. Automatic microaneurysm detection using the sparse principal component analysis-based unsupervised classification method. IEEE Access 2017, 5, 2563–2572. [Google Scholar] [CrossRef]
- Omar, M.; Khelifi, F.; Tahir, M.A. Detection and classification of retinal fundus images exudates using region based multiscale LBP texture approach. In Proceedings of the 2016 International Conference on Control, Decision and Information Technologies (CoDIT), Saint Julian’s, Malta, 6–8 April 2016. [Google Scholar]
- Vijayan, T.; Sangeetha, M.; Kumaravel, A.; Karthik, B. Gabor filter and machine learning based diabetic retinopathy analysis and detection. Microprocess. Microsyst. 2020, 103353. [Google Scholar] [CrossRef]
- Ishtiaq, U.; Abdul Kareem, S.; Abdullah, E.R.M.F.; Mujtaba, G.; Jahangir, R.; Ghafoor, H.Y. Diabetic retinopathy detection through artificial intelligent techniques: A review and open issues. Multimed. Tools Appl. 2020, 79, 15209–15252. [Google Scholar] [CrossRef]
- Foundation Consumer Healthcare. EyePACS: Diabetic Retinopathy Detection. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection/data (accessed on 13 February 2023).
- Mafarja, M.M.; Eleyan, D.; Jaber, I.; Hammouri, A.; Mirjalili, S. Binary dragonfly algorithm for feature selection. In Proceedings of the 2017 International conference on New Trends in Computing Sciences (ICTCS), Amman, Jordan, 11–13 October 2017. [Google Scholar]
- Mirjalili, S. SCA: A sine cosine algorithm for solving optimization problems. Knowl. Based Syst. 2016, 96, 120–133. [Google Scholar] [CrossRef]
- Mubarak, D. Classification of early stages of esophageal cancer using transfer learning. IRBM 2022, 43, 251–258. [Google Scholar]
- Akram, M.U.; Khalid, S.; Tariq, A.; Khan, S.A.; Azam, F. Detection and classification of retinal lesions for grading of diabetic retinopathy. Comput. Biol. Med. 2014, 45, 161–171. [Google Scholar] [CrossRef]
- Luo, Y.; Pan, J.; Fan, S.; Du, Z.; Zhang, G. Retinal image classification by self-supervised fuzzy clustering network. IEEE Access 2020, 8, 92352–92362. [Google Scholar] [CrossRef]
- Wang, J.; Bai, Y.; Xia, B. Simultaneous diagnosis of severity and features of diabetic retinopathy in fundus photography using deep learning. IEEE J. Biomed. Health Inform. 2020, 24, 3397–3407. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.A.; Laude, A.; Faye, I.; Tang, T.B. Automated microaneurysm detection in diabetic retinopathy using curvelet transform. J. Biomed. Opt. 2016, 21, 101404. [Google Scholar] [CrossRef] [PubMed]
- Orlando, J.I.; Prokofyeva, E.; Del Fresno, M.; Blaschko, M.B. An ensemble deep learning based approach for red lesion detection in fundus images. Comput. Methods Programs Biomed. 2018, 153, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, C.; Jain, S.; Sood, M. Hierarchical severity grade classification of non-proliferative diabetic retinopathy. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 2649–2670. [Google Scholar] [CrossRef]
- Lam, C.; Yu, C.; Huang, L.; Rubin, D. Retinal lesion detection with deep learning using image patches. Investig. Ophthalmol. Vis. Sci. 2018, 59, 590–596. [Google Scholar] [CrossRef]
- Keerthiveena, B.; Esakkirajan, S.; Subudhi, B.N.; Veerakumar, T. A hybrid BPSO-SVM for feature selection and classification of ocular health. IET Image Process. 2021, 15, 542–555. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Wang, X.H.; Wang, X.F.; Shih, F.Y. Retinal vessels segmentation based on level set and region growing. Pattern Recognit. 2014, 47, 2437–2446. [Google Scholar] [CrossRef]
- Ali, A.; Qadri, S.; Khan Mashwani, W.; Kumam, W.; Kumam, P.; Naeem, S.; Goktas, A.; Jamal, F.; Chesneau, C.; Anam, S. Machine learning based automated segmentation and hybrid feature analysis for diabetic retinopathy classification using fundus image. Entropy 2020, 22, 567. [Google Scholar] [CrossRef]
- Majumder, S.; Kehtarnavaz, N. Multitasking deep learning model for detection of five stages of diabetic retinopathy. IEEE Access 2021, 9, 123220–123230. [Google Scholar] [CrossRef]
- Li, Y.-H.; Yeh, N.-N.; Chen, S.-J.; Chung, Y.-C. Computer-assisted diagnosis for diabetic retinopathy based on fundus images using deep convolutional neural network. Mob. Inf. Syst. 2019, 2019, 6142839. [Google Scholar] [CrossRef]
- Jabbar, M.K.; Yan, J.; Xu, H.; Ur Rehman, Z.; Jabbar, A. Transfer Learning-Based Model for Diabetic Retinopathy Diagnosis Using Retinal Images. Brain Sci. 2022, 12, 535. [Google Scholar] [CrossRef] [PubMed]
- Bilal, A.; Zhu, L.; Deng, A.; Lu, H.; Wu, N. AI-Based Automatic Detection and Classification of Diabetic Retinopathy Using U-Net and Deep Learning. Symmetry 2022, 14, 1427. [Google Scholar] [CrossRef]
- Luo, X.; Wang, W.; Xu, Y.; Lai, Z.; Jin, X.; Zhang, B.; Zhang, D. A deep convolutional neural network for diabetic retinopathy detection via mining local and long-range dependence. CAAI Trans. Intell. Technol. 2023. [Google Scholar] [CrossRef]







| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| SVM | Linear | 0 | 98.01 | 93.10 | 99.43 | 99.87 | 96.16 |
| 1 | 98.22 | 98.84 | 99.71 | 98.53 | |||
| 2 | 99.73 | 98.97 | 99.74 | 99.35 | |||
| 3 | 99.33 | 98.68 | 99.67 | 99.00 | |||
| 4 | 99.63 | 94.38 | 98.52 | 96.94 | |||
| Quadratic | 0 | 98.63 | 94.27 | 99.72 | 99.93 | 96.92 | |
| 1 | 99.77 | 99.20 | 99.80 | 99.48 | |||
| 2 | 99.90 | 99.24 | 99.81 | 99.57 | |||
| 3 | 99.50 | 99.97 | 99.99 | 99.73 | |||
| 4 | 99.70 | 95.22 | 98.75 | 97.41 | |||
| Fine Gaussian | 0 | 41.70 | 94.33 | 43.21 | 69.00 | 59.27 | |
| 1 | 6.630 | 7.480 | 79.48 | 7.030 | |||
| 2 | 4.630 | 5.490 | 80.06 | 5.030 | |||
| 3 | 54.07 | 100.00 | 100.00 | 70.19 | |||
| 4 | 48.83 | 89.66 | 98.59 | 63.23 | |||
| Medium Gaussian | 0 | 94.77 | 92.87 | 99.18 | 99.81 | 95.92 | |
| 1 | 90.87 | 90.26 | 97.55 | 90.56 | |||
| 2 | 91.07 | 90.52 | 97.62 | 90.79 | |||
| 3 | 99.27 | 99.93 | 99.98 | 99.60 | |||
| 4 | 99.77 | 94.33 | 98.50 | 96.97 | |||
| Coarse Gaussian | 0 | 95.68 | 91.17 | 98.10 | 99.56 | 94.51 | |
| 1 | 92.90 | 95.71 | 98.96 | 94.28 | |||
| 2 | 97.90 | 91.32 | 97.68 | 94.50 | |||
| 3 | 96.90 | 99.97 | 99.99 | 98.41 | |||
| 4 | 99.53 | 94.02 | 98.42 | 96.70 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| KNN | Fine | 0 | 64.43 | 91.47 | 92.83 | 98.23 | 92.14 |
| 1 | 18.90 | 18.57 | 79.28 | 18.73 | |||
| 2 | 18.60 | 18.53 | 79.55 | 18.56 | |||
| 3 | 99.40 | 99.87 | 99.97 | 99.63 | |||
| 4 | 93.80 | 94.02 | 98.51 | 93.91 | |||
| Medium | 0 | 95.75 | 96.73 | 99.62 | 99.91 | 98.16 | |
| 1 | 90.03 | 95.14 | 98.85 | 92.52 | |||
| 2 | 96.80 | 98.91 | 99.73 | 97.84 | |||
| 3 | 98.70 | 93.41 | 98.26 | 95.98 | |||
| 4 | 96.47 | 92.11 | 97.93 | 94.24 | |||
| Coarse | 0 | 76.45 | 88.53 | 98.19 | 99.59 | 93.11 | |
| 1 | 48.57 | 47.93 | 86.81 | 48.25 | |||
| 2 | 52.23 | 47.57 | 85.61 | 49.79 | |||
| 3 | 94.57 | 100.00 | 100.00 | 97.21 | |||
| 4 | 98.37 | 94.46 | 98.56 | 96.37 | |||
| Cosine | 0 | 74.75 | 92.43 | 96.12 | 99.07 | 94.24 | |
| 1 | 63.87 | 44.38 | 79.99 | 52.37 | |||
| 2 | 18.20 | 34.89 | 91.51 | 23.92 | |||
| 3 | 99.30 | 99.37 | 99.84 | 99.33 | |||
| 4 | 99.93 | 92.67 | 98.03 | 96.17 | |||
| Weighted | 0 | 62.51 | 91.07 | 94.30 | 98.63 | 92.66 | |
| 1 | 13.90 | 13.61 | 77.94 | 13.75 | |||
| 2 | 13.97 | 13.72 | 78.03 | 13.84 | |||
| 3 | 98.87 | 100.00 | 100.00 | 99.43 | |||
| 4 | 94.73 | 94.17 | 98.53 | 94.45 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| SVM | Linear | 0 | 98.17 | 92.80 | 99.07 | 99.78 | 95.83 |
| 1 | 99.43 | 98.58 | 99.64 | 99.00 | |||
| 2 | 99.57 | 99.20 | 99.80 | 99.38 | |||
| 3 | 99.33 | 99.97 | 99.99 | 99.65 | |||
| 4 | 99.73 | 94.33 | 98.50 | 96.95 | |||
| Quadratic | 0 | 98.35 | 93.10 | 99.54 | 99.89 | 96.21 | |
| 1 | 99.67 | 98.91 | 99.73 | 99.29 | |||
| 2 | 99.73 | 99.30 | 99.83 | 99.52 | |||
| 3 | 99.47 | 99.97 | 99.99 | 99.72 | |||
| 4 | 99.77 | 94.33 | 98.50 | 96.97 | |||
| Fine Gaussian | 0 | 43.61 | 85.57 | 50.82 | 79.30 | 63.77 | |
| 1 | 5.100 | 5.830 | 79.40 | 5.440 | |||
| 2 | 3.530 | 4.210 | 79.92 | 3.840 | |||
| 3 | 46.83 | 100.00 | 100.00 | 63.79 | |||
| 4 | 77.03 | 67.91 | 90.90 | 72.18 | |||
| Medium Gaussian | 0 | 92.86 | 92.87 | 99.15 | 99.80 | 95.90 | |
| 1 | 86.20 | 85.54 | 96.36 | 85.87 | |||
| 2 | 86.17 | 85.88 | 96.46 | 86.02 | |||
| 3 | 99.30 | 99.93 | 99.98 | 99.62 | |||
| 4 | 99.77 | 94.24 | 98.48 | 96.92 | |||
| Coarse Gaussian | 0 | 85.23 | 91.97 | 97.87 | 99.50 | 94.83 | |
| 1 | 47.17 | 80.58 | 97.16 | 59.50 | |||
| 2 | 89.70 | 62.35 | 86.46 | 73.56 | |||
| 3 | 97.67 | 100.00 | 100.00 | 98.82 | |||
| 4 | 99.63 | 94.02 | 98.42 | 96.75 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| KNN | Fine | 0 | 64.14 | 92.03 | 92.50 | 98.13 | 92.26 |
| 1 | 18.03 | 17.79 | 79.17 | 17.91 | |||
| 2 | 17.70 | 17.66 | 79.38 | 17.68 | |||
| 3 | 99.63 | 99.90 | 99.98 | 99.77 | |||
| 4 | 93.30 | 94.05 | 98.53 | 93.67 | |||
| Medium | 0 | 75.57 | 91.57 | 97.83 | 99.49 | 94.59 | |
| 1 | 64.13 | 43.75 | 79.38 | 52.01 | |||
| 2 | 18.57 | 33.64 | 90.84 | 23.93 | |||
| 3 | 99.03 | 100.00 | 100.00 | 99.51 | |||
| 4 | 99.57 | 94.32 | 98.50 | 96.87 | |||
| Coarse | 0 | 76.46 | 89.37 | 98.13 | 99.58 | 93.55 | |
| 1 | 47.97 | 47.04 | 86.50 | 47.50 | |||
| 2 | 50.13 | 47.18 | 85.97 | 48.61 | |||
| 3 | 96.23 | 100.00 | 100.00 | 98.08 | |||
| 4 | 98.60 | 94.38 | 98.53 | 96.45 | |||
| Cosine | 0 | 74.74 | 93.00 | 95.65 | 98.94 | 94.30 | |
| 1 | 64.33 | 44.41 | 79.87 | 52.55 | |||
| 2 | 16.87 | 33.96 | 91.80 | 22.54 | |||
| 3 | 99.57 | 98.97 | 99.74 | 99.27 | |||
| 4 | 99.93 | 92.85 | 98.08 | 96.26 | |||
| Weighted | 0 | 62.45 | 91.57 | 94.50 | 98.67 | 93.01 | |
| 1 | 13.77 | 13.44 | 77.83 | 13.60 | |||
| 2 | 12.67 | 12.62 | 78.07 | 12.64 | |||
| 3 | 99.27 | 100.00 | 100.00 | 99.63 | |||
| 4 | 94.97 | 94.03 | 98.49 | 94.49 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| SVM | Linear | 0 | 98.29 | 93.07 | 99.36 | 99.85 | 96.11 |
| 1 | 99.57 | 98.81 | 99.70 | 99.19 | |||
| 2 | 99.67 | 99.17 | 99.79 | 99.42 | |||
| 3 | 99.43 | 99.97 | 99.99 | 99.70 | |||
| 4 | 99.70 | 94.41 | 98.53 | 96.98 | |||
| Quadratic | 0 | 98.85 | 95.23 | 99.86 | 99.97 | 97.49 | |
| 1 | 99.87 | 99.37 | 99.84 | 99.62 | |||
| 2 | 99.93 | 99.27 | 99.82 | 99.60 | |||
| 3 | 99.47 | 99.97 | 99.99 | 99.72 | |||
| 4 | 99.77 | 95.96 | 98.95 | 97.83 | |||
| Fine Gaussian | 0 | 41.19 | 94.23 | 45.92 | 72.26 | 61.75 | |
| 1 | 4.200 | 4.470 | 77.56 | 4.330 | |||
| 2 | 2.400 | 2.670 | 78.09 | 2.530 | |||
| 3 | 56.33 | 100.00 | 100.00 | 72.07 | |||
| 4 | 48.77 | 89.53 | 98.58 | 63.14 | |||
| Medium Gaussian | 0 | 95.01 | 93.07 | 99.43 | 99.87 | 96.14 | |
| 1 | 91.47 | 90.56 | 97.62 | 91.01 | |||
| 2 | 91.37 | 91.00 | 97.74 | 91.18 | |||
| 3 | 99.40 | 100.00 | 100.00 | 99.70 | |||
| 4 | 99.73 | 94.44 | 98.53 | 97.02 | |||
| Coarse Gaussian | 0 | 96.85 | 91.23 | 98.03 | 99.54 | 94.51 | |
| 1 | 97.30 | 96.82 | 99.20 | 97.06 | |||
| 2 | 98.93 | 95.74 | 98.90 | 97.31 | |||
| 3 | 97.23 | 99.93 | 99.98 | 98.56 | |||
| 4 | 99.57 | 94.11 | 98.44 | 96.76 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| KNN | Fine | 0 | 61.66 | 91.87 | 93.20 | 98.33 | 92.53 |
| 1 | 12.03 | 11.81 | 77.48 | 11.92 | |||
| 2 | 11.33 | 11.30 | 77.78 | 11.32 | |||
| 3 | 99.53 | 99.83 | 99.96 | 99.68 | |||
| 4 | 93.70 | 94.04 | 98.52 | 93.87 | |||
| Medium | 0 | 74.07 | 91.23 | 98.03 | 99.54 | 94.51 | |
| 1 | 69.67 | 43.34 | 77.23 | 53.44 | |||
| 2 | 11.20 | 26.71 | 92.32 | 15.78 | |||
| 3 | 98.63 | 100.00 | 100.00 | 99.31 | |||
| 4 | 99.60 | 94.29 | 98.49 | 96.87 | |||
| Coarse | 0 | 77.11 | 88.90 | 98.16 | 99.58 | 93.30 | |
| 1 | 50.03 | 49.25 | 87.10 | 49.64 | |||
| 2 | 52.97 | 48.85 | 86.14 | 50.82 | |||
| 3 | 95.07 | 100.00 | 100.00 | 97.47 | |||
| 4 | 98.60 | 94.50 | 98.57 | 96.51 | |||
| Cosine | 0 | 74.54 | 92.70 | 96.30 | 99.11 | 94.46 | |
| 1 | 69.97 | 44.39 | 78.08 | 54.31 | |||
| 2 | 10.50 | 27.75 | 93.17 | 15.24 | |||
| 3 | 99.53 | 99.24 | 99.81 | 99.38 | |||
| 4 | 100.00 | 92.62 | 98.01 | 96.17 | |||
| Weighted | 0 | 60.66 | 91.27 | 94.28 | 98.62 | 92.75 | |
| 1 | 9.830 | 9.520 | 76.63 | 9.670 | |||
| 2 | 8.800 | 8.750 | 77.06 | 8.780 | |||
| 3 | 98.97 | 100.00 | 100.00 | 99.48 | |||
| 4 | 94.43 | 94.09 | 98.52 | 94.26 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| SVM | Linear | 0 | 98.20 | 92.80 | 99.07 | 99.78 | 95.83 |
| 1 | 99.47 | 98.61 | 99.65 | 99.04 | |||
| 2 | 99.53 | 99.27 | 99.82 | 99.40 | |||
| 3 | 99.43 | 99.97 | 99.99 | 99.70 | |||
| 4 | 99.73 | 94.33 | 98.50 | 96.95 | |||
| Quadratic | 0 | 98.41 | 93.23 | 99.50 | 99.88 | 96.27 | |
| 1 | 99.67 | 99.07 | 99.77 | 99.37 | |||
| 2 | 99.77 | 99.40 | 99.85 | 99.58 | |||
| 3 | 99.57 | 100.00 | 100.00 | 99.78 | |||
| 4 | 99.80 | 94.36 | 98.51 | 97.00 | |||
| Fine Gaussian | 0 | 42.95 | 90.33 | 51.83 | 79.35 | 65.86 | |
| 1 | 2.910 | 3.320 | 77.46 | 3.100 | |||
| 2 | 2.000 | 2.060 | 76.58 | 2.030 | |||
| 3 | 47.07 | 100.00 | 100.00 | 64.01 | |||
| 4 | 75.10 | 79.22 | 95.16 | 77.10 | |||
| Medium Gaussian | 0 | 92.88 | 92.17 | 99.35 | 99.85 | 95.63 | |
| 1 | 86.44 | 85.16 | 96.23 | 85.79 | |||
| 2 | 86.67 | 86.24 | 96.54 | 86.45 | |||
| 3 | 99.37 | 99.93 | 99.98 | 99.65 | |||
| 4 | 99.77 | 94.30 | 98.49 | 96.95 | |||
| Coarse Gaussian | 0 | 85.05 | 92.17 | 97.81 | 99.48 | 94.90 | |
| 1 | 41.20 | 86.43 | 98.38 | 55.80 | |||
| 2 | 94.33 | 61.19 | 85.04 | 74.23 | |||
| 3 | 97.77 | 99.90 | 99.98 | 98.82 | |||
| 4 | 99.77 | 94.06 | 98.43 | 96.83 | |||
| CLASSIFIER | CLASS | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | |
|---|---|---|---|---|---|---|---|
| KNN | Fine | 0 | 61.36 | 92.53 | 93.63 | 98.43 | 93.08 |
| 1 | 10.43 | 10.30 | 77.28 | 10.37 | |||
| 2 | 10.27 | 10.23 | 77.48 | 10.25 | |||
| 3 | 99.73 | 99.90 | 99.98 | 99.82 | |||
| 4 | 93.83 | 94.12 | 98.53 | 93.97 | |||
| Medium | 0 | 74.31 | 91.73 | 97.73 | 99.47 | 94.64 | |
| 1 | 70.07 | 43.66 | 77.40 | 53.80 | |||
| 2 | 10.83 | 26.62 | 92.53 | 15.40 | |||
| 3 | 99.27 | 99.93 | 99.98 | 99.60 | |||
| 4 | 99.63 | 94.32 | 98.50 | 96.90 | |||
| Coarse | 0 | 76.28 | 89.67 | 98.14 | 99.58 | 93.71 | |
| 1 | 47.13 | 46.41 | 86.39 | 46.77 | |||
| 2 | 49.17 | 46.50 | 85.86 | 47.80 | |||
| 3 | 96.60 | 100.00 | 100.00 | 98.27 | |||
| 4 | 98.83 | 94.37 | 98.53 | 96.55 | |||
| Cosine | 0 | 74.23 | 93.10 | 95.88 | 99.00 | 94.47 | |
| 1 | 68.70 | 43.94 | 78.08 | 53.60 | |||
| 2 | 9.73 | 25.50 | 92.89 | 14.09 | |||
| 3 | 99.60 | 98.81 | 99.70 | 99.20 | |||
| 4 | 100.00 | 92.97 | 98.11 | 96.35 | |||
| Weighted | 0 | 60.36 | 91.70 | 94.08 | 98.56 | 92.88 | |
| 1 | 8.40 | 8.200 | 76.49 | 8.300 | |||
| 2 | 7.90 | 7.870 | 76.88 | 7.890 | |||
| 3 | 99.43 | 99.93 | 99.98 | 99.68 | |||
| 4 | 94.33 | 94.11 | 98.53 | 94.22 | |||
| Ref. | Year | No. of Classes | Performance Measures | ||||
|---|---|---|---|---|---|---|---|
| ACC (%) | SEN (%) | SPE (%) | PRE (%) | F1 (%) | |||
| [35] | 2021 | 5 | 82.00 | 64.00 | - | 69.00 | 66.00 |
| [36] | 2019 | 5 | 86.17 | 89.30 | 90.89 | - | - |
| [37] | 2022 | 5 | 96.61 | 94.90 | 98.40 | 98.50 | 96.70 |
| [38] | 2022 | 5 | 97.92 | 96.94 | 97.44 | 96.90 | 97.10 |
| [39] | 2023 | 5 | 83.60 | 86.50 | 69.30 | 81.90 | 82.60 |
| Proposed | 5 | 98.85 | 98.85 | 99.71 | 98.89 | 98.85 | |
| CLASSIFIER | 5-Fold Cross-Validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | ||||||||||||
| ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | Time (s) | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | Time (s) | ||
| SVM | Linear | 98.01 | 98.00 | 98.06 | 99.50 | 98.00 | 81.62 | 98.17 | 98.17 | 98.23 | 99.54 | 98.17 | 190.21 |
| Quadratic | 98.63 | 98.63 | 98.67 | 99.66 | 98.62 | 101.13 | 98.35 | 98.35 | 98.41 | 99.59 | 98.34 | 251.35 | |
| Fine Gaussian | 41.70 | 41.70 | 49.17 | 85.43 | 40.95 | 1337.80 | 43.61 | 43.61 | 45.75 | 85.90 | 41.81 | 2722.50 | |
| Medium Gaussian | 94.77 | 94.77 | 94.85 | 98.69 | 94.77 | 183.54 | 92.86 | 92.86 | 92.95 | 98.22 | 92.87 | 439.25 | |
| Coarse Gaussian | 95.68 | 95.68 | 95.82 | 98.92 | 95.68 | 212.74 | 85.23 | 85.23 | 86.97 | 96.31 | 84.69 | 431.17 | |
| KNN | Fine | 64.43 | 64.43 | 64.76 | 91.11 | 64.60 | 223.19 | 64.14 | 64.14 | 64.38 | 91.04 | 64.26 | 432.57 |
| Medium | 95.75 | 95.75 | 95.84 | 98.94 | 95.75 | 223.35 | 74.57 | 74.57 | 73.91 | 93.64 | 73.38 | 431.72 | |
| Coarse | 76.45 | 76.45 | 77.63 | 94.11 | 76.95 | 223.89 | 76.46 | 76.46 | 77.35 | 94.12 | 76.84 | 432.51 | |
| Cosine | 74.75 | 74.75 | 73.49 | 93.69 | 73.21 | 225.20 | 74.74 | 74.74 | 73.17 | 93.69 | 72.98 | 440.06 | |
| Weighted | 62.51 | 62.51 | 63.16 | 90.63 | 62.83 | 246.57 | 62.45 | 62.45 | 62.92 | 90.61 | 62.68 | 434.06 | |
| CLASSIFIER | 10-Fold Cross-Validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 3 | Experiment 4 | ||||||||||||
| ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | Time (s) | ACC (%) | SEN (%) | PRE (%) | SPE (%) | F1 (%) | Time (s) | ||
| SVM | Linear | 98.29 | 98.29 | 98.34 | 99.57 | 98.28 | 169.26 | 98.20 | 98.19 | 98.25 | 99.55 | 98.19 | 418.31 |
| Quadratic | 98.85 | 98.85 | 98.89 | 99.71 | 98.85 | 180.88 | 98.41 | 98.41 | 98.47 | 99.60 | 98.40 | 470.49 | |
| Fine Gaussian | 41.19 | 41.19 | 48.52 | 85.30 | 40.76 | 2042.40 | 42.95 | 43.48 | 47.29 | 85.71 | 42.42 | 5534.27 | |
| Medium Gaussian | 95.01 | 95.01 | 95.09 | 98.75 | 95.01 | 301.35 | 92.88 | 92.88 | 93.00 | 98.22 | 92.89 | 799.76 | |
| Coarse Gaussian | 96.85 | 96.85 | 96.93 | 99.21 | 96.84 | 346.30 | 85.05 | 85.05 | 87.88 | 96.26 | 84.12 | 793.14 | |
| KNN | Fine | 61.66 | 61.69 | 62.04 | 90.41 | 61.86 | 270.30 | 61.36 | 61.36 | 61.63 | 90.34 | 61.50 | 1087.80 |
| Medium | 74.07 | 74.07 | 72.47 | 93.52 | 71.98 | 273.56 | 74.31 | 74.31 | 72.45 | 93.58 | 72.07 | 1759.80 | |
| Coarse | 77.11 | 77.11 | 78.15 | 94.28 | 77.55 | 257.45 | 76.28 | 76.28 | 77.08 | 94.07 | 76.62 | 1557.70 | |
| Cosine | 74.54 | 74.54 | 72.06 | 93.64 | 71.91 | 252.98 | 74.23 | 74.23 | 71.42 | 93.56 | 71.54 | 1545.30 | |
| Weighted | 60.66 | 60.66 | 61.33 | 90.17 | 60.99 | 247.85 | 60.36 | 60.35 | 60.84 | 90.09 | 60.59 | 480.97 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishtiaq, U.; Abdullah, E.R.M.F.; Ishtiaque, Z. A Hybrid Technique for Diabetic Retinopathy Detection Based on Ensemble-Optimized CNN and Texture Features. Diagnostics 2023, 13, 1816. https://doi.org/10.3390/diagnostics13101816
Ishtiaq U, Abdullah ERMF, Ishtiaque Z. A Hybrid Technique for Diabetic Retinopathy Detection Based on Ensemble-Optimized CNN and Texture Features. Diagnostics. 2023; 13(10):1816. https://doi.org/10.3390/diagnostics13101816
Chicago/Turabian StyleIshtiaq, Uzair, Erma Rahayu Mohd Faizal Abdullah, and Zubair Ishtiaque. 2023. "A Hybrid Technique for Diabetic Retinopathy Detection Based on Ensemble-Optimized CNN and Texture Features" Diagnostics 13, no. 10: 1816. https://doi.org/10.3390/diagnostics13101816






