Investigation of Eye-Tracking Scan Path as a Biomarker for Autism Screening Using Machine Learning Algorithms
Abstract
:1. Introduction
2. Literature Review
3. Materials and Methods
3.1. Data
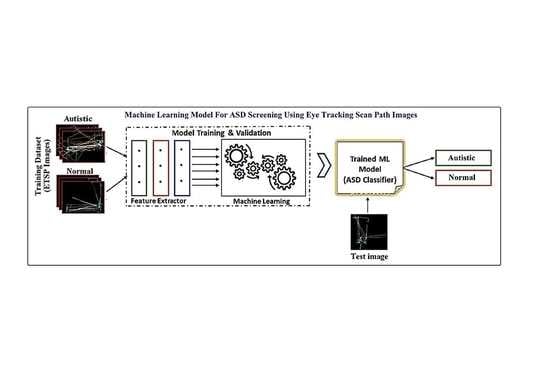
3.2. Overview of the Proposed ML Model
- (a)
- Pre-Processing
- (b)
- Feature Extraction by PCA
- (c)
- Classical ML Classifier
- (d)
- Feature Extraction by CNN
- (e)
- DNN Classifier
- (f)
- Building the ML Models
3.3. Experimentations and Results
- Step 1: Conversion of ETSP images to grayscale images and resizing
- Step 2: Application of PCA to Extract Features
- Step 3: Creation of ML Model
- Step 4: Design of DNN Models
- Step 5: Creation of artificially populated more extensive dataset
- Step 6: Experimentations on the augmented dataset
4. Discussion
4.1. Clinical Implications
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bölte, S.; Hallmayer, J. FAQs on Autism, Asperger Syndrome, and Atypical Autism Answered by International Experts; Hogrefe Publishing: Cambridge, MA, USA, 2010. [Google Scholar]
- CDC. Screening and Diagnosis of Autism Spectrum Disorder for Healthcare Providers. Available online: https://www.cdc.gov/ncbddd/autism/hcp-screening.html (accessed on 27 January 2022).
- Maenner, M.J.; Shaw, K.A.; Baio, J. Prevalence of Autism Spectrum Disorder among Children Aged 8 Years; CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Dawson, G. Early behavioral intervention, brain plasticity, and the prevention of autism spectrum disorder. Dev. Psychopathol. 2008, 20, 775–803. [Google Scholar] [CrossRef] [PubMed]
- APA (American Psychological Association). Anxiety disorders. In Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; APA: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Zwaigenbaum, L.; Penner, M. Autism spectrum disorder—Advances in diagnosis and evaluation. BMJ 2018, 361, k1674. [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.S.; Novak, M.M.; Zubritsky, C.D. Factors associated with age of diagnosis among children with autism spectrum disorder. Pediatrics 2005, 116, 1480–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Jiang, M.; Duchesne, X.M.; Laugeson, E.A.; Kennedy, D.P.; Adolphs, R.; Zhao, Q. Atypical Visual Saliency in Autism Spectrum Disorder Quantified through Model-Based Eye Tracking. Neuron 2015, 88, 604–616. [Google Scholar] [CrossRef] [Green Version]
- Pierce, K.; Marinero, S.; Hazin, R.; McKenna, B.; Barnes, C.C.; Malige, A. Eye Tracking Reveals Abnormal Visual Preference for Geometric Images as an Early Biomarker of an Autism Spectrum Disorder Subtype Associated With Increased Symptom Severity. Biol. Psychiatry 2016, 79, 657–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkmer, T.; Anderson, K.; Falkmer, M.; Horlin, C. Diagnostic procedures in autism spectrum disorders: A systematic literature review. Eur. Child Adolesc. Psychiatry 2013, 22, 329–340. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Al-Shamma, O.; Fadhel, M.A.; Farhan, L.; Zhang, J. Classification of Red Blood Cells in Sickle Cell Anemia Using Deep Convolutional Neural Network. Intell. Syst. Des. App. 2020, 940, 550–559. [Google Scholar]
- Rubin, J.; Parvaneh, S.; Rahman, A.; Conroy, B.; Babaeizadeh, S. Densely connected convolutional networks for detection of atrial fibrillation from short single-lead ECG recordings. J. Electrocardiol. 2018, 1, 518–521. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Mujeeb Rahman, K.K.; Subashini, M.M. A Deep Neural Network-Based Model for Screening Autism Spectrum Disorder Using the Quantitative Checklist for Autism in Toddlers (QCHAT). J. Autism. Dev. Disord. 2021. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Mujeeb Rahman, K.K.; Subashini, M.M. Identification of Autism in Children Using Static Facial Features and Deep Neural Networks. Brain Sci. 2022, 12, 94. [Google Scholar] [CrossRef] [PubMed]
- Koppe, G.; Meyer-Lindenberg, A.; Durstewitz, D. Deep learning for small and big data in psychiatry. Neuropsychopharmacology 2021, 46, 176–190. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, L.; Zhang, Z.; He, H. Algorithm Research on Improving Activation Function of Convolutional Neural Networks. In Proceedings of the Chinese Control and Decision Conference (CCDC), Nanchang, China, 3–5 June 2019; pp. 3582–3586. [Google Scholar]
- Dhillon, A.; Verma, G.K. Convolutional neural network: A review of models, methodologies and applications to object detection. Prog. Artif. Intell. 2020, 9, 85–112. [Google Scholar] [CrossRef]
- Almourad, M.B.; Bataineh, E.; Stocker, J.; Marir, F. Analyzing the Behavior of Autistic and Normal Developing Children Using Eye Tracking Data. In Proceedings of the International Conference on Kansei Engineering & Emotion Research, Kuching, Malaysia, 19–22 March 2018. [Google Scholar]
- Duan, H.; Zhai, G.; Min, X.; Che, Z.; Fang, Y.; Yang, X.; Callet, P.L. A dataset of eye movements for the children with autism spectrum disorder. In Proceedings of the 10th ACM Multimedia Systems Conference, Amherst, MA, USA, 18–21 June 2019; pp. 255–260. [Google Scholar]
- Tao, Y.; Shyu, M. SP-ASDNet: CNN-LSTM Based ASD Classification Model using Observer ScanPaths. In Proceedings of the IEEE International Conference on Multimedia & Expo Workshops (ICMEW), Shanghai, China, 8–12 July 2019. [Google Scholar]
- Carette, R.; Cilia, F.; Dequen, G.; Bosche, J.; Guerin, J.L.; Vandromme, L. Automatic Autism Spectrum Disorder Detection Thanks to Eye-Tracking and Neural Network-Based Approach. In Internet of Things (IoT) Technologies for HealthCare. HealthyIoT 2017. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering; Ahmed, M., Begum, S., Fasquel, J.B., Eds.; Springer: Cham, Swiztherland, 2018; Volume 225. [Google Scholar] [CrossRef]
- Carette, R.; Elbattah, M.; Dequen, G.; Guérin, J.-L.; Cilia, F. Visualization of Eye-Tracking Patterns in Autism Spectrum Disorder Method and Dataset. In Proceedings of the Thirteenth International Conference on Digital Information Management (ICDIM), Berlin, Germany, 24–26 September 2018; pp. 248–253. [Google Scholar]
- Carette, R.; Elbattah, M.; Cilia, F.; Dequen, G.; Guérin, J.; Bosche, J. Learning to Predict Autism Spectrum Disorder based on the Visual Patterns of Eye-tracking Scanpaths. In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies—HEALTHINF, Prague, Czech Republic, 22–24 February 2019; pp. 103–4305. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Franco, F.O.; Revers, M.C.; Silva, A.F.; Portolese, J.; Brentani, H.; Machado-Lima, A.; Nunes, F.L.S. Computer-aided autism diagnosis based on visual attention models using eye tracking. Sci. Rep. 2021, 11, 10131. [Google Scholar] [CrossRef]
- Elbattah, M. Visualization of Eye-Tracking Scan path in Autism Spectrum Disorder: Image Dataset. In Proceedings of the 12th International Conference on Health Informatics, Prague, Czech Republic, 22–24 February 2019. [Google Scholar] [CrossRef]
- Wang, X.; Paliwal, K.K. Feature extraction and dimensionality reduction algorithms and their applications in vowel recognition. Pattern Recognit. 2003, 36, 2429–2439. [Google Scholar] [CrossRef]
- Hess, A.S.; Hess, J.R. Principal component analysis. Transfusion 2018, 58, 1580–1582. [Google Scholar] [CrossRef]
- Coadou, Y. Boosted Decision Trees and Applications. Eur. Phys. J. Conf. 2013, 55, 02004. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Wiering, M.A.; Schutten, M.; Millea, A.; Meijster, A.; Schomaker, L.R.B. Deep Support Vector Machines for Regression Problems; Institute of Artificial Intelligence and Cognitive Engineering, University of Groningen: Groningen, The Netherlands, 2013. [Google Scholar]
- Shotton, J.; Sharp, T.; Kohli, P.; Nowozin, S.; Winn, J.; Criminisi, A. Decision Jungles: Compact and Rich Models for Classification; Microsoft Research: Redmond, WA, USA, 2015. [Google Scholar]
- Quiterio, T.M.; Lorena, A.C. Determining the Structure of Decision Directed Acyclic Graphs for Multiclass Classification Problems. In Proceedings of the 2016 5th Brazilian Conference on Intelligent Systems (BRACIS), Recife, Brazil, 9–12 October 2016; pp. 115–120. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Xin, M.; Wang, Y. Research on image classification model based on deep convolution neural network. EURASIP J. Image Video Process. 2019, 2019, 40. [Google Scholar] [CrossRef] [Green Version]
- Indolia, S.; Goswami, A.K.; Mishra, S.P.; Asopa, P. Conceptual Understanding of Convolutional Neural Network—A Deep Learning Approach. Procedia Comput. Sci. 2018, 132, 679–688. [Google Scholar] [CrossRef]
- Agatonovic Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Bengio, Y. Learning Deep Architectures for AI. Found. Trends® Mach. Learn. 2009, 21. [Google Scholar]
- Uzair, M.; Jamil, N. Effects of Hidden Layers on the Efficiency of Neural networks. In Proceedings of the 23rd International Multitopic Conference (INMIC), Bahawalpur, Pakistan, 5–7 November 2020. [Google Scholar]
- Yao, G.; Lei, T.; Zhong, J. A review of convolutional-neural-network-based action recognition. Pattern Recogn Lett. 2019, 118, 14–22. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Mikolajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018. [Google Scholar]
- Florkowski, C.M. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: Communicating the performance of diagnostic tests. Clin. Biochem. Rev. 2008, 29, S83–S87. [Google Scholar]
- Browne, M.W. Cross-Validation Methods. J. Math. Psychol. 1999, 44, 108–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekaba, B. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; A Comprehensive Guide for Beginners; Apress: Berkeley, CA, USA, 2019; ISBN 978-1-4842-4470-8. [Google Scholar] [CrossRef]
- Donati, G.; Davis, R.; Forrester, G.S. Gaze behaviour to lateral face stimuli in infants who do and do not receive an ASD diagnosis. Sci. Rep. 2020, 10, 13185. [Google Scholar]
- Klin, A.; Lin, D.J.; Gorrindo, P.; Ramsay, G.; Jones, W. Two-year-olds with autism orient to nonsocial contingencies rather than biological motion. Nature 2009, 459, 257–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, J.P.; Bedford, R.; Jones, E.J.; Charman, T.; Johnson, M.H.; Gliga, T. Gaze Following and Attention to Objects in Infants at Familial Risk for ASD. Front. Psychol. 2019, 10, 1799. [Google Scholar] [CrossRef] [Green Version]
- Klin, A.; Jones, W.; Schultz, R.; Volkmar, F.; Cohen, D. Visual fixation patterns during viewing of naturalistic social situations as predictors of social competence in individuals with autism. Arch. Gen. Psychiatry 2002, 59, 809–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feczko, E.; Miranda-Dominguez, O.; Marr, M.; Graham, A.M.; Nigg, J.T.; Fair, D.A. The Heterogeneity Problem: Approaches to Identify Psychiatric Subtypes. Trends Cogn. Sci. 2019, 23, 584–601. [Google Scholar] [CrossRef] [PubMed]
- Lenroot, R.K.; Yeung, P.K. Heterogeneity within Autism Spectrum Disorders: What have We Learned from Neuroimaging Studies? Front. Hum. Neurosci. 2013, 7, 733. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Description | Details |
|---|---|
| Number of participants | 59 |
| Number of classes | 2 |
| Number of TD | 29 |
| Number of ASD—Diagnosed | 30 |
| Total number of ETSP images | 547 |
| ETSP Images—TD | 328 |
| ETSP Images—ASD | 219 |
| Size of image | 640 × 480 × 3 |
| CARS Scores (Mean) | 32.97 |
| Age in years and Mean age | 3–13, 7.88 |
| % Gender distribution (M:F) | 64:36 |
| Dataset | Total Number of Images | Number of TD. Images | Number of ASD. Images | Size of the Image |
|---|---|---|---|---|
| Dataset I (Original) | 547 | 328 | 219 | 640 × 480 × 3 |
| Dataset II (Augmented dataset) | 2566 | 1519 | 1041 | 640 × 480 × 3 |
| Dataset | Model | %Sensitivity | %Specificity | %PPV | %NPV | %AUC |
|---|---|---|---|---|---|---|
| Original | DSVM | 44.06 | 55.55 | 52.0 | 47.61 | 51.50 |
| DJ | 55.93 | 64.80 | 63.46 | 57.37 | 60.40 | |
| BDT | 64.40 | 57.40 | 62.20 | 59.60 | 67.60 | |
| DNN | 78.57 | 75.47 | 87.12 | 62.50 | 78.00 |
| Layer ID | Type/Function | Dimensions (Number of Kernel and Size) | Dimensions of Output Feature |
|---|---|---|---|
| Level 1 | 2D convolution Activation-ReLU | 16, 3 × 3 | 16, 100 × 100 |
| Max pooling | 16, 2 × 2 | 16, 50 × 50 | |
| Level 2 | 2D convolution Activation-ReLU | 32, 3 × 3 | 32, 50 × 50 |
| Max pooling | 32, 2 × 2 | 32, 25 × 25 | |
| Level 3 | 2D convolution Activation-ReLU | 64, 3 × 3 | 64, 25 × 25 |
| Max pooling | 64, 2 × 2 | 64, 12 × 12 | |
| Level 4 | 2D convolution Activation-ReLU | 128, 3 × 3 | 128, 12 × 12 |
| Max pooling | 128, 2 × 2 | 128, 6 × 6 | |
| Flatten Layer | - | 1 × 4608 | |
| Dense Layer 1 Dense Layer 2 | 256 128 | - | |
| Output Layer (Sigmoid Activation) | 1 | - |
| Dataset | Model | %Sensitivity | %Specificity | %PPV | %NPV | %AUC |
|---|---|---|---|---|---|---|
| Original | DSVM | 43.90 | 72.80 | 51.51 | 66.60 | 61.60 |
| DJ | 38.28 | 84.15 | 61.05 | 67.75 | 66.80 | |
| BDT | 49.80 | 86.50 | 70.56 | 72.66 | 74.60 | |
| DNN | 93.28 | 91.38 | 94.46 | 90.06 | 97.00 |
| Dataset | Model | %Sensitivity | %Specificity | %PPV | %NPV | %AUC |
|---|---|---|---|---|---|---|
| Mini batch | DNN | 58.62 | 86.67 | 80.95 | 64.00 | 72.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanhirakadavath, M.R.; Chandran, M.S.M. Investigation of Eye-Tracking Scan Path as a Biomarker for Autism Screening Using Machine Learning Algorithms. Diagnostics 2022, 12, 518. https://doi.org/10.3390/diagnostics12020518
Kanhirakadavath MR, Chandran MSM. Investigation of Eye-Tracking Scan Path as a Biomarker for Autism Screening Using Machine Learning Algorithms. Diagnostics. 2022; 12(2):518. https://doi.org/10.3390/diagnostics12020518
Chicago/Turabian StyleKanhirakadavath, Mujeeb Rahman, and Monica Subashini Mohan Chandran. 2022. "Investigation of Eye-Tracking Scan Path as a Biomarker for Autism Screening Using Machine Learning Algorithms" Diagnostics 12, no. 2: 518. https://doi.org/10.3390/diagnostics12020518