1. Introduction
China is a modern agricultural country supplying fruit products, wherein the fruit planting area is relatively large. Due to its rich nutritional and medicinal value, the apple has become one of China’s four major fruits. However, diseases in apple leaves cause major production and economic losses, as well as reductions in both the quality and quantity of the fruit industry output. Apple leaf disease detection has received increasing attention for the monitoring of large apple orchards.
Traditionally, plant disease severity is scored with visual inspection of plant tissues by trained experts [
1], which leads to expensive cost and low efficiency. With the popularization of digital cameras and the advance of information technology in agriculture, cultivation and management expert systems have been widely used, greatly improving the production capacity of plants [
2]. However, for the “expert system”, extraction and expression characteristics of pests and diseases mainly depend on expert experience, which easily leads to a relative lack of standardization and low recognition rates. With the popularity of machine learning algorithms in computer vision, in order to improve the accuracy and rapidity of the diagnosis results, researchers have studied automated plant disease diagnosis based on traditional machine learning algorithms, such as random forest,
k-nearest neighbor, and Support Vector Machine (SVM) [
3,
4,
5,
6,
7,
8,
9,
10,
11,
12]. However, because the classification features are selected and adopted based on human experience, these approaches improved the recognition accuracy, but the recognition rate is still not high enough and is vulnerable to artificial feature selection. Developed in recent years, the deep convolutional neutral network approach is an end-to-end pipeline that can automatically discover the discriminative features for image classification, whose advantages lie in the use of shared weights to reduce the memory footprint and improve performance, and the direct input of the image into the model. Until now, the convolutional neural network has been regarded as one of the best classification approaches for pattern recognition tasks. Inspired by the breakthrough of the convolutional neutral network in image-based recognition, the use of convolutional neural networks to identify early disease images has become a new research hotspot in agricultural informatization. In [
13,
14,
15,
16,
17,
18,
19,
20], convolutional neural networks (CNNs) are widely studied and used in the field of crop disease recognition. These studies show that convolutional neural networks have not only reduced the demand of image preprocessing, but also improved the recognition accuracy.
In this paper, we present a novel identifying approach for apple leaf diseases based on a deep convolutional neural network. The CNN-based approach faces two difficulties. First of all, apple pathological images are not sufficient for the training model. Second, determining the best structures of the network model is fundamentally a more difficult task.
The main contributions of this paper are summarized as follows:
In order to solve the problem of insufficient apple pathological images, this paper proposes a training image generation technology based on image processing techniques, which can enhance the robustness and prevent overfitting of the CNN-based model in the training process. Natural apple pathological images are first acquired and are then processed in order to generate sufficient pathological images using digital image processing technologies such as image rotation, brightness adjustment, and PCA (Principal Component Analysis) jittering to disturb natural images; these are able to simulate the real environment of image acquisition, and expanding the pathological images gives an important guarantee of generalization capability of the convolutional neural network model.
A convolutional neural network is first employed to diagnose apple leaf diseases; the end-to-end learning model can automatically discover the discriminative features of the apple pathological images and identify the four common types of apple leaf diseases with high accuracy. By analyzing the characteristics of apple leaf diseases, a novel deep convolutional neural network model based on AlexNet is proposed; the convolution kernel size is adjusted, fully-connected layers are replaced by a convolutional layer, and GoogLeNet’s Inception is applied to improve the feature extraction ability.
The experimental results show that the proposed CNN-based model achieves an accuracy of 97.62% on the hold-out test set, which is higher than the other traditional models. Compared with the standard AlexNet model, the parameters of the proposed model are significantly decreased by 51,206,928, demonstrating the faster convergence rate. Using the dataset of 13,689 synthetic images of diseased apple leaves, the identification rate increases by 10.83% over that of the original natural images, proving the better generalization ability and robustness.
The remainder of this paper is organized as follows. In
Section 2, related work is introduced and summarized. In
Section 3, based on apple leaf pathological image acquisition and image processing technology, sufficient training images are generated.
Section 4 describes the novel deep convolutional neural network model.
Section 5 analyzes the experimental results provided by the identification approach to apple leaf diseases based on CNNs. Finally, this paper is concluded in
Section 6.
2. Related Work
Plant diseases are a major threat to production and quality, and many researchers have made various efforts to control these diseases. In the last few years, traditional machine learning algorithms have been widely used to realize disease detection. In [
6], Qin et al. proposed a feasible solution for lesion image segmentation and image recognition of alfalfa leaf disease. The ReliefF method was first used to extract a total of 129 features, and then an SVM model was trained with the most important features. The results indicated that image recognition of the four alfalfa leaf diseases can be implemented and obtained an average accuracy of 94.74%. In [
7], Rothe et al. presented a pattern recognition system for identifying and classifying three cotton leaf diseases. Using the captured dataset of natural images, an active contour model was used for image segmentation and Hu’s moments were extracted as features for the training of an adaptive neuro-fuzzy inference system. The pattern recognition system achieved an average accuracy of 85%. In [
8], Islam et al. presented an approach that integrated image processing and machine learning to allow the diagnosis of diseases from leaf images. This automated method classifies diseases on potato plants from ‘Plant Village’, which is a publicly available plant image database. The segmentation approach and utilization of an SVM demonstrated disease classification in over 300 images, and obtained an average accuracy of 95%. In [
9], Gupta proposed an autonomously modified SVM-CS (Cuckoo Search) model to identify the healthy portion and disease. Using a dataset of diseases containing plant leaves suffering from Alternaria Alternata, Cercospora Leaf Spot, Anthracnose, and Bacterial Blight, along with healthy leaf images, the proposed model was trained and optimized using the concept of a cuckoo search. However, identification and classification approaches of these studies are semiautomatic and complex, and deal with a series of image processing technologies. At the same time, it is very difficult to accurately detect the specific disease images without extracting and designing the appropriate classification features depending heavily on expert experience.
Recently, several researchers have studied plant disease identification based on deep learning approaches. In [
16], Lu et al. proposed a novel identification approach for rice diseases based on deep convolutional neural networks. Using a dataset of 500 natural images of diseased and healthy rice leaves and stems, CNNs were trained to identify 10 common rice diseases. The experimental results showed that the proposed model achieved an average accuracy of 95.48%. In [
17], Tan et al. presented an approach based on CNN to recognize apple pathologic images, and employed a self-adaptive momentum rule to update CNN parameters. The results demonstrated that the recognition accuracy of the proposal was up to 96.08%, with a fairly quick convergence. In [
18], a novel cucumber leaf disease detection system was presented based on convolutional neural networks. Under the fourfold cross-validation strategy, the proposed CNN-based system achieved an average accuracy of 94.9% in classifying cucumbers into two typical disease classes and a healthy class. The experimental results indicate that a CNN-based model can automatically extract the requisite classification features and obtain the optimal performance. In [
14], Sladojevic et al. proposed a novel approach based on deep convolutional networks to detect plant disease. By discriminating the plant leaves from their surroundings, 13 common different types of plant diseases were recognized by the proposed CNN-based model. The experimental results showed that the proposed CNN-based model can reach a good recognition performance, and obtained an average accuracy of 96.3%. In [
19], Mohanty et al. developed a CNN-based model to detect 26 diseases and 14 crop species. Using a public dataset of 54,306 images of diseased and healthy plant leaves, the proposed model was trained and achieved an accuracy of 99.35%. These studies show that convolution neural networks have been widely applied to the field of crop and plant disease recognition, and have obtained good results. However, on the one hand, these studies only apply the CNN-based models to identify crop and plant diseases without improving the model. On the other hand, so far, the CNN-based model has not been applied to the identification of apple leaf diseases; a novel CNN-based model developed by our research group is applied to detect apple leaf diseases in this paper.
3. Generating Apple Pathological Training Images
3.1. Apple Leaf Pathological Image Acquisition
Appropriate datasets are required at all stages of object recognition research, from training the CNN-based models to evaluating the performance of the recognition algorithms [
14].
In this section, four common apple leaf diseases were chosen as the research objects, whose lesions are more widespread than others and do great harm to apple quality and quantity. A total of 1053 images with typical disease symptoms were acquired, consisting of 252 images of Mosaic (caused by Papaya ringspot virus), 319 images of Brown spot (caused by Marssonina coronaria), 182 images of Rust (caused by Pucciniaceae glue rust), and 300 images of Alternaria leaf spot (caused by Alternaria alternaria f.sp mali). Images of apple leaf diseases were supplied by two apple experiment stations, which are in Qingyang county, Gansu Province, China and Baishui county, Shanxi Province, China. A BM-500GE/BB-500GE digital color camera was used to capture apple leaf disease images with resolutions of 2456 × 2058 pixels. After processing using digital image processing technology, the number of images was expanded in order to train the proposed model.
Figure 1 shows that the difference among the four apple leaf diseases is obvious. Firstly, the lesions caused by the same disease show a certain commonality under similar natural conditions. Secondly, the yellow lesion of Mosaic diffuses throughout the leaf, which is different from the other disease lesions. The aforementioned observations contribute to the diagnosis and recognition of the diseases. However, the similarity between Rust and Alternaria leaf spot in terms of geometric features increases the complexity of distinguishing the apple leaf diseases. Finally, the lesion of Brown Spot is brown with an irregular edge of green, which is different from the others and relatively easy to detect.
3.2. Image Processing and Generating Pathological Images
The overfitting problem of deep learning models appears when a statistical model describes random noise or errors rather than the underlying relationship [
21]. In order to reduce overfitting at the training stage and enhance the anti-interference ability of complex conditions, a slight distortion is introduced to the images at the experimental stage.
3.2.1. Direction Disturbance
In the apple orchard, the relative position of the image acquisition device to the research object is determined by their current spatial relation, which depends on the shooting position [
17]. Therefore, it is difficult to photograph apple leaf pathological images from every angle to meet all the possibilities. In this section, for testing and constructing the adaptability of the CNN-based model, an expanded image dataset is established from the natural images using rotation transformation and mirror symmetry.
Image rotation occurs when all pixels rotate a certain angle around the center of the image. Assume that is an arbitrary point of the image; after rotating ° counterclockwise, the point’s coordinate is . The coordinates of the calculation of two points are shown in Equations (1) and (2).
The horizontal mirror symmetry takes a vertical line in an image as the axis, and all pixels of the image are exchanged. Assume that represents the width, and that an arbitrary point’s coordinate is ; after mirror symmetry, the point’s coordinate is .
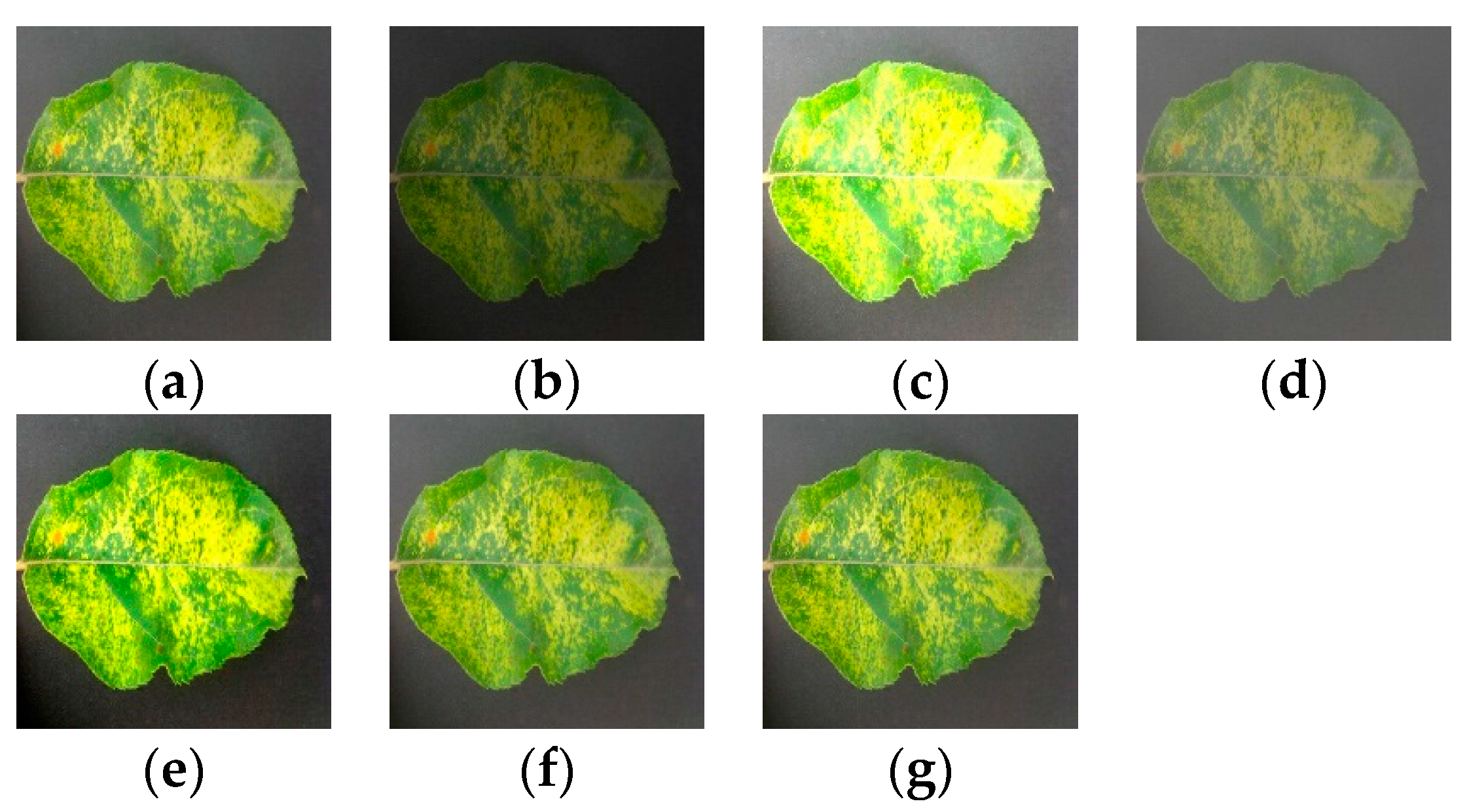
As shown in
Figure 2, a pathological image is rotated and mirrored to generate four pathological images, in which the angle of rotation consists of 90 °, 180 °and 270 °, and mirror symmetry includes horizontal symmetry.
3.2.2. Light Disturbance
The light condition often becomes complex during image collection, owing to the interference of many factors, especially weather factors [
17]—variable sunlight orientation, the random occurrence of cloud, the disturbance of sand and dust, hazy weather, etc. These factors probably influence the brightness and balance of acquired images. To improve the generalization ability of the learning model, it must be trained with expanded leaf disease images that imitate different light backgrounds. Based on an original image, six apple leaf pathological images are generated by adjusting the sharpness value, brightness value, and contrast value.
Image sharpening can enhance image edges and borders to make the object emerge from the picture. Assuming an RGB image pixel
, the Laplacian template is applied into the image using Equation (3):
For the alteration of image brightness, the RGB value of pixels needs to be increased or decreased randomly. Assume that
represents the original RGB value,
is the adjusted value, and
represents the brightness transformation factor. The formula is as follows:
For the contrast of the image, the larger RGB value is increased and the smaller RGB value is reduced, based on the median value of the brightness. The formula is as follows:
where
represents the median value of the brightness, and the other parameters have the same meaning as in Equation (4).
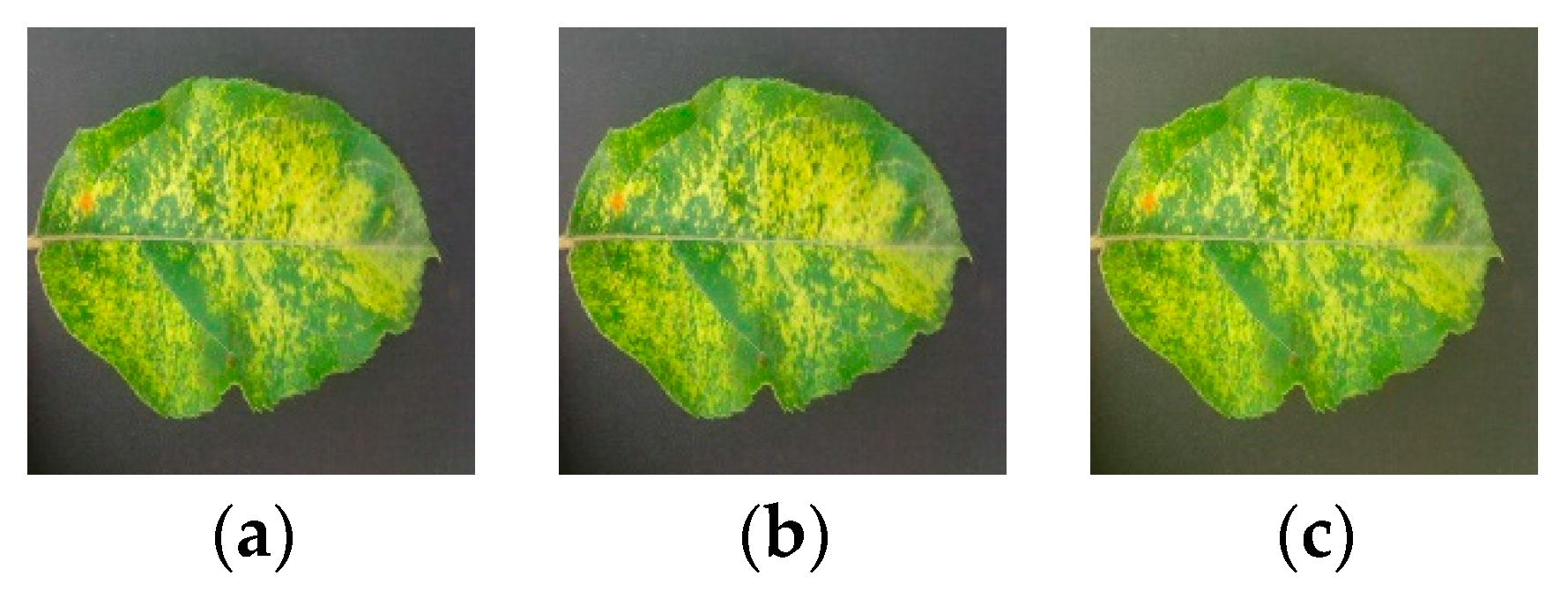
In addition to the direction disturbance and light disturbance, Gaussian noise and PCA jittering are also employed on the original apple leaf pathological images.
The original images are disturbed by Gaussian noise, which can simulate the possible noise caused by equipment in the image acquisition process. First, random numbers are generated consistent with a Gaussian distribution. Then, the random numbers are added to the original pixel values of the image, which finally compresses the sums to the [0, 255] interval.
PCA jittering was proposed by Alex et al. [
22], and is used to reduce overfitting. In this paper, it is applied to expand the dataset. To each RGB image pixel
, the following quantity is added:
where
and
are the
ith feature vector and eigenvalue of the 3 × 3 covariance matrix of RGB pixel values, respectively, and
is the random variable.
The light disturbance is illustrated in
Figure 3, with the six pathological images generated by adjusting the brightness, contrast, and sharpness.
Figure 4 visualizes the Gaussian noise and PCA jittering against the pathological image.
For this stage, 12 pathological natural images can be derived from an original apple image. Finally, an image database containing 10,888 images for training and 2801 images for testing was created. Chosen from experience, the size of the images was compressed from 2456 × 2058 to 256 × 256, which is able to be divided by 2 and reduces the training time [
16].
4. Building the Deep Convolutional Neural Network
Inspired by the classical AlexNet [
22], GoogLeNet [
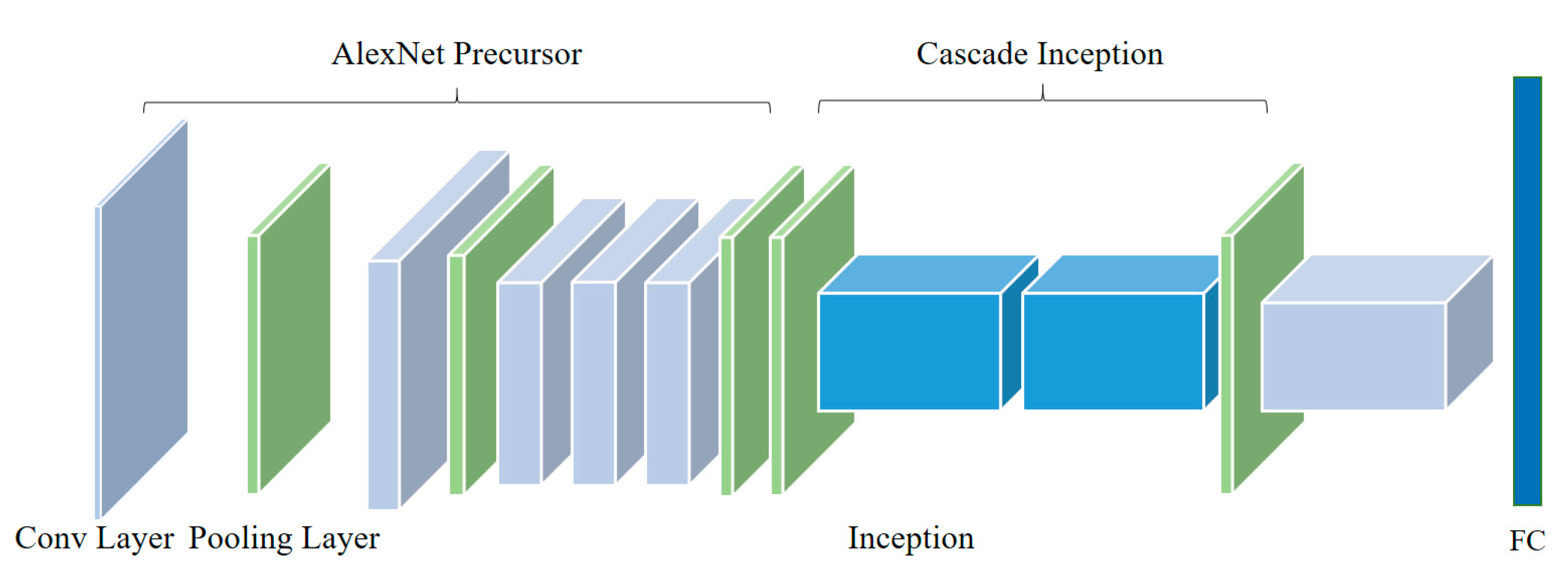
23], and their performance improvements, a deep convolutional neural network model is proposed to identify apple leaf diseases. The proposed CNN-based model and related parameters are shown in
Figure 5 and
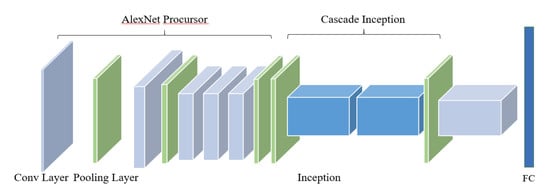
Table 1. First of all, a structure named AlexNet Precursor is designed, which is based on the standard AlexNet model. For the perception of the convolution kernel, a larger sized the convolution kernel has a stronger ability to extract the macro information of the image, and vice versa. A lesion is smaller than the whole image, and other information on the image can be understood as “noise” which needs to be filtered. As a consequence, the first convolutional layer is designed to be 96 kernels of size 9 × 9 × 3, which is different from the first convolutional layer’s kernel size of 11 × 11 × 3 in the standard AlexNet. The second convolutional layer filters the noise with 256 kernels of size 5 × 5 × 48; response-normalization layers follow the first two convolutional layers, which are themselves followed by max-pooling layers. The third convolutional layer has 384 kernels with a size of 3 × 3 × 256 connected to the (normalized, pooled) outputs of the second convolutional layer. The fourth layer is filtered with 384 kernels of size 3 × 3 × 192, and the fifth layer has 256 kernels with a size of 2 × 2 × 192 to improve the ability to extract small features, which is also different from the standard AlexNet, and is then followed by a max-pooling layer.
After AlexNet Precursor, an architecture named Cascade Inception is designed including two max-pooling layers and two Inception structures. The first max-pooling layer is applied to filter the noise of feature maps generated by AlexNet Precursor, and the two Inceptions then extract the optimal discrimination features from multidimension analysis. Feature maps before the first Inception are input into the second Inception’s concatenation layer, which prevents some of the features being filtered by these two Inceptions. Meanwhile, the sixth convolutional layer followed by the Cascade Inception has 4096 kernels with a size of 1 × 1 × 736, which replaces the first two fully connected layers of the standard AlexNet. The fully connected layer is adjusted to predict four classes of apple leaf diseases, and the final layer is a four-way Softmax layer.
More specifically, the convolution layer, pooling layer, activation function, and Softmax layer in the novel CNN-based model are described below.
4.1. Convolution Layer
The output feature map of each convolution layer is determined by a convolution operation between the upper feature maps of the current layer and convolution kernels. Generally, the output feature map could be indicated by Equation (7):
where
means the
layer,
represents the convolutional kernel,
is the bias, and
is a set of input feature maps [
16].
4.2. Max-Pooling Layer
The max-pooling layer, which is a form of nonlinear down-sampling, could reduce the size of the feature maps gained from the convolutional layers to achieve spatial invariance, which leads to faster convergence and improves the generalization performance [
24].
When the feature map
is passed to the max-pooling layer, the max operation is applied to the feature map
, which produces a pooled feature map
as the output. As shown in Equation (8), the max operation selects the largest element:
where
represents pooling region
in feature map
, and
is the index of each element within it;
denotes the pooled feature maps [
25].
4.3. Softmax Regression
Softmax regression is used for multiclassification problems. The hypothesis function is shown in Equation (9):
The model parameters
are trained to minimize the cost function
. In the equation below, 1{.} is the indicator function, so that 1{a true statement} = 1, and 1{a false statement} = 0. The cost function
is shown in Equation (10):
The training database is denoted ,. In Softmax regression, the possibility of classifying into category is
4.4. ReLU Activation Function
The activation function determines the neural network data processing method, and influences the learning ability of the neural network model. The ReLU activation function has a fast convergence speed and alleviates the problem of overfitting. As a result, this method is used for the output of every convolutional layer. The ReLU activation function formula is shown in Equation (12).
4.5. GoogLeNet’s Inception
A special structure named Inception is the main feature of GoogLeNet; it keeps the sparse network structure, and utilizes an intensive matrix of high-performance computing. As shown in
Figure 6, the Inception consists of parallel 1 × 1, 3 × 3, and 5 × 5 convolutional layers as well as a max-pooling layer to extract a variety of features in parallel. Then, 1 × 1 convolution layers are added for dimensionality reduction. Finally, a filter concatenation layer concatenates simply the output of all these parallel layers [
23].
4.6. Nesterov’s Accelerated Gradient (NAG)
The training process of convolutional neural networks includes two stages of a feedforward pass and a backpropagation pass. In the backpropagation pass stage, the error is passed from higher layers to lower layers.
Stochastic Gradient Descent (SGD) is used to update the weight for convolutional neural networks. However, SGD may lead to the “local optimum” problem. To solve this problem, Nesterov’s Accelerated Gradient (NAG) is applied to train the proposed CNN-based model. As a convex optimization algorithm, NAG has a higher rate of convergence. The updated weights are calculated based on the last iteration, as shown in the Equations (13) and (14):
where
represents the current update vector,
represents the last update vector,
is the current updated parameter,
represents
’s gradient in the objective function,
is the momentum term, and
represents the learning rate [
26].
5. Experimental Evaluation
In this section, the experimental setup is first introduced, and details of the experimental platform and benchmarks are provided. Finally, experimental results are analyzed and discussed.
5.1. Experimental Setup
The experiment was performed on an Ubuntu workstation equipped with an Intel(R) Xeon(R) CPU E5-2650 v4 @ 2.20 GHz, accelerated by two NVIDIA Tesla P100 GPUs. The NVIDIA Tesla P100 has 3584 CUDA cores, and 16 GB of HBM2 memory. The core frequency is up to 1328 MHz and the floating-point performance is 10.6 TFLOPS. The CNN-based model was implemented in the Caffe deep learning framework [
27]. More detailed configuration parameters are presented in
Table 2.
This paper uses the four common types of apple leaf diseases to evaluate the novel CNN-based model. These apple pathological images were collected in Qingyang County, Gansu Province, China and Baishui County, Shanxi Province, China. After application of image processing techniques, the generated pathological images constituted a dataset of 13,689 images of diseased apple leaves; the numbers of various pathological images in the training and test sets are presented in
Table 3.
5.2. Accuracy and Learning Convergence Comparison
In this section, other learning models such as SVM and BP neural networks, standard AlexNet, GoogLeNet, ResNet-20, VGGNet-16, and the proposed model are trained on the expanded dataset. ResNet-20, AlexNet, and GoogLeNet were trained over 40 epochs with a learning rate of 0.01, and SGD was chosen as the optimization algorithm. The proposed model was trained using the NAG optimization algorithm with a learning rate of 0.001. In addition, VGGNet-16 was conducted by transfer learning, with a learning rate of 0.0001. As shown in
Table 4, because the adjustment of the convolutional layers is based on the features of apple leaf disease images, the experimental results show that the proposed model achieved an accuracy of 97.62% on the testing set, which is higher than that of other models. In addition, the AlexNet model has a good recognition ability and obtains an average accuracy of 91.19%. GoogLeNet has multiple Inceptions and possesses the ability for multidimensional feature extraction; however, its network is not adjusted by features of apple pathological images, and a final recognition rate of 95.69% is realized. ResNet-20, as a residual neural network, obtains an accuracy of 92.76%. VGGNet-16 realized a recognition rate of 96.32% with transfer learning. In addition, the SVM model with the SGD optimizer and BP neural networks obtained an accuracy of 68.73% and 54.63%, respectively. The experimental results show that the traditional approaches rely heavily on classification features designed by experts to enhance recognition accuracy, while the level of expert experience has a significant influence on the selection of classification features. Compared with the traditional approaches, the CNN-based approaches could not only automatically extract the best classification features from multiple dimensions, but also learn layered features, from low-level features, such as edge, corner, and color, to high-level semantic features, such as shape and object, to improve the recognition performance on apple leaf diseases.
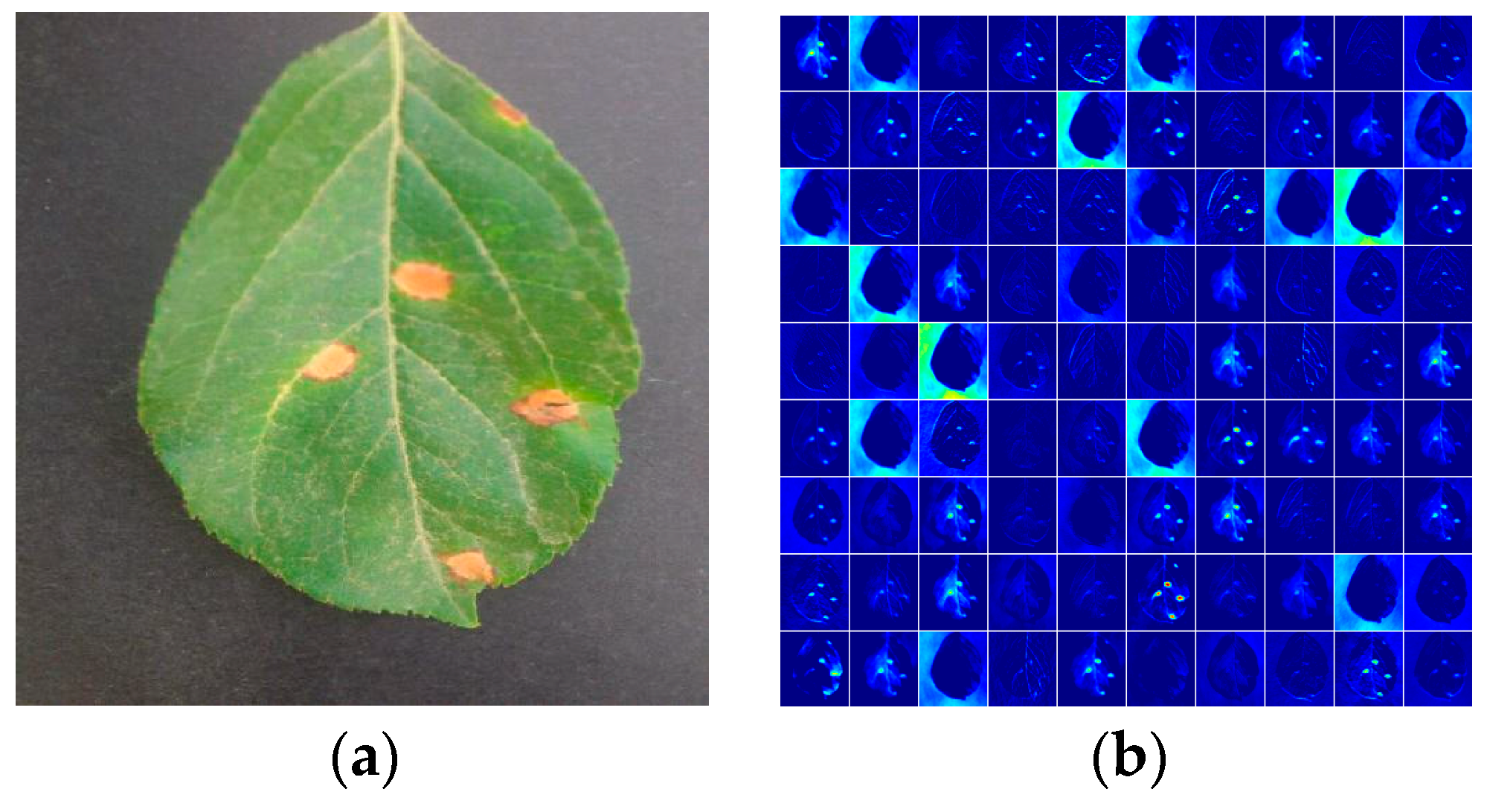
Table 5 shows the confusion matrix of our model, and the fraction of accurately predicted images for each of the four apple leaf diseases is presented in detail. As the analysis of the four diseases in the above section showed, the characteristics of Mosaic and Brown spot are very different from the others, and recognition rates of 100.00% and 99.29 were achieved for Mosaic and Brown spot, respectively. However, Alternaria leaf spot is extremely similar to Rust in geometric features, which leads to their lower recognition rates. As shown in
Figure 7, pathological features in the original image are extracted by the proposed model with GoogLeNet Inception, which improves the automatic feature extraction in a multidimensional space. Hence, the proposed CNN-based model has a better identification ability with regards to apple leaf diseases.
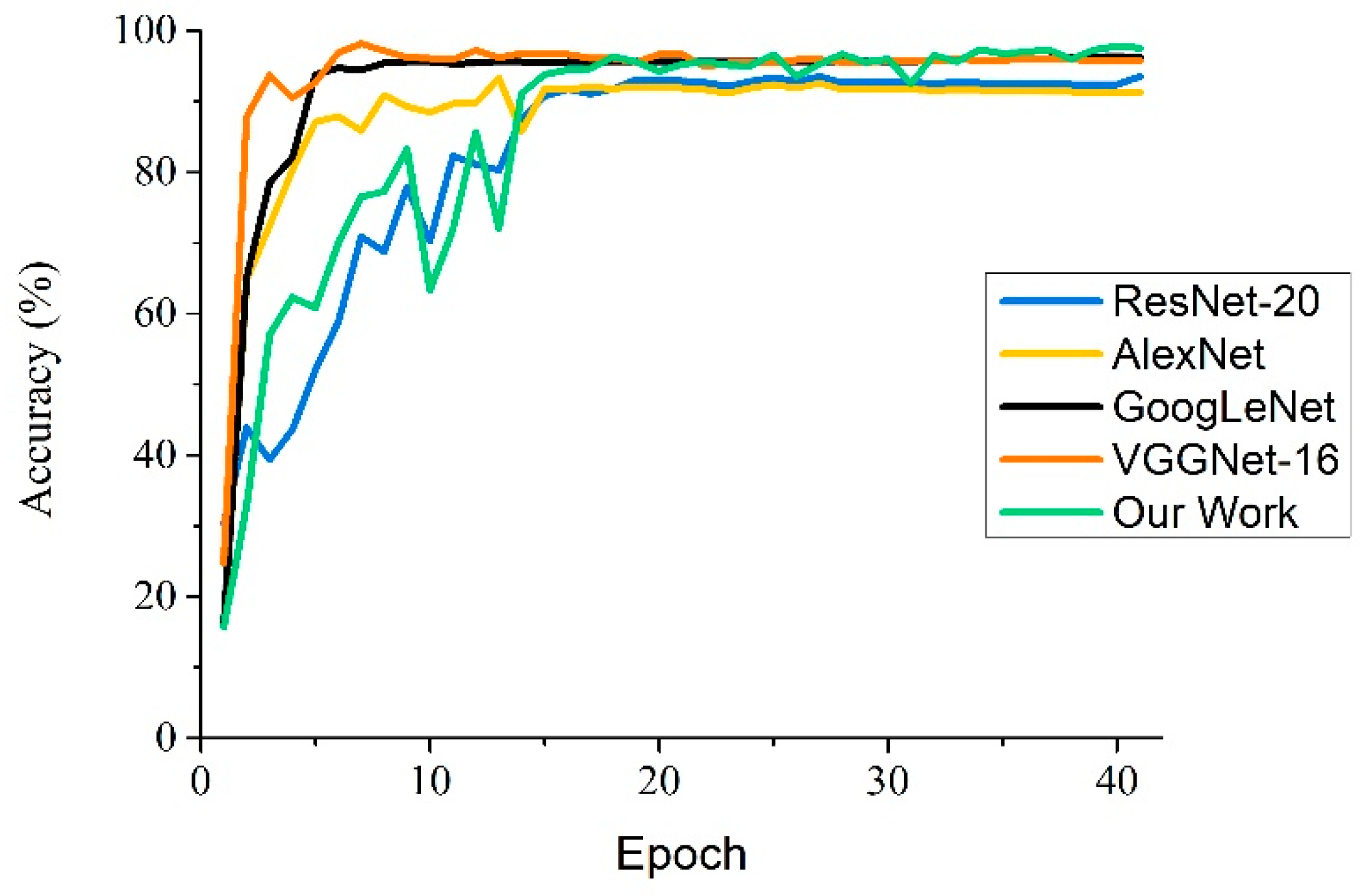
In addition, in this experiment, the five CNN-based models were selected to research the variation of accuracy with the training epochs. As shown in
Figure 8, the four classical convolutional neural networks and the proposed model begin to converge after a certain number of epochs and finally achieve their optimal recognition performance. On the whole, the training processes of GoogLeNet, VGGNet-16, and AlexNet are basically stable after 10 epochs, and other models have a satisfactory convergence after 15 epochs. Because of the use of transfer learning, VGGNet-16 has a faster convergence speed than other CNN-based models, and achieves an accuracy of 96.32%. Because GoogLeNet uses Inception structures—which have a strong ability for feature learning—to extract the features of apple leaf diseases, the convergence point of GoogLeNet occurs at 10 epochs. Compared to other neutral networks, AlexNet uses a traditional network structures, which results in slower convergence—the starting point of convergence is at about 16 epochs. As for ResNet-20, the strategy of batch normalization improves its convergence rate, and the model reaches convergence at 20 epochs. In our work, Inception structures, removing the partial fully connected layers, and the NAG optimization algorithm are used for the proposed model; compared with the standard AlexNet, the proposed model improves the convergence speed of the network model, begins to converge at about 14 epochs, and provides higher recognition accuracy for apple leaf diseases.
In addition, to prevent overfitting in this paper, various methods were performed. First, various digital image processing technologies such as image rotation, mirror symmetry, brightness adjustment, and PCA jittering, were applied to the natural training images to simulate the real acquisition environment and increase the diversity and quantity of the apple pathological training images, which can prevent the overfitting problem and make the proposed model generalize better during the training process. Second, the response-normalization layers were used in the proposed model to achieve local normalization, which is thought of as an effective way to prevent the overfitting problem. Third, by replacing some of the fully connected layers with convolution layers, the proposed model has fewer training parameters than the standard CNN-based model, and this scheme aids the generation of the model.
5.3. Computational Resources
In computational theory, the simplest computational resources are computation time, the number of parameters necessary to solve a problem, and memory space [
28]. In this section, the computational resource comparisons of four classic neural network models and the proposed model are analyzed in
Table 6. Compared with other learning models, although the proposed model is trained with batch size 128, it takes the least video memory space for training. The standard AlexNet has the minimum training time among all the CNN-based models. Compared with AlexNet, the proposed model not only has a similar training time, but also achieves a higher accuracy of recognition. As for ResNet-20, it has the least learned weights, but takes up a great deal of memory space and takes the longest time to train parameters. Overall, the proposed model uses less computational resources to build the model and acquires the best accuracy in identifying apple leaf diseases, which allows it to meet the needs of real production.
5.4. The Effect of Pooling Layers for Identifying Leaf Diseases
In order to verify the influence of the inserted max-pooling layers—that is, the first layer in the structure of the Cascade Inception—on the identification accuracy, a contrast experiment was performed under the same experimental conditions.
The experiment showed that the novel CNN-based model with inserted max-pooling layers achieves an accuracy of 97.62%, and the proposed model without inserted max-pooling layers only obtains an accuracy of 93.29%. The experimental result shows that the recognition accuracy is improved by about 4%, which is because pooling layers can filter the noise in feature maps, which can cause the Inception structures to better extract features and thus improve the recognition accuracy.
5.5. The Effect of Optimization Algorithms for Identification Accuracy
The optimization algorithm is also important for the performance of the recognition rate. In this section, the SGD optimization algorithm with a learning rate of 0.01 and the NAG optimization algorithm with a learning rate of 0.001 are used to train the CNN-based model; the learning rate of the NAG algorithm is stepped down by 80% every 10 epochs, whose learning rates are parameters of their own best performance.
As shown in
Figure 9, the CNN-based model with SGD achieves an accuracy of 93.32%, while an accuracy of 97.62% is obtained by the model with the NAG optimizer. The phenomenon indicates that the model based on the SGD optimizer has a “local optimum” problem. The SGD optimizer updates the parameters based on the current batch and the current position, which causes the update direction to be very unstable. In general, the negative gradient direction is used as the forward direction. This is because this direction is the fastest descent direction from the current position. However, if the target function is a nonconvex optimization, the SGD optimizer tends to fall into the “local optimum”. While the NAG optimizer updates the parameters, it is not only influenced by the previous update, but also uses the current batch gradient to fine-tune the final direction, which improves the stability of the training process and has the ability to overcome the “local optimum” problem.
At the same time, as shown in
Figure 9, the result shows that the training process of the proposed model almost converged after 25 epochs, and finally achieved an accuracy of 97.62%. The reason for this phenomenon is that the learning rate decreases gradually to almost the invariant, which greatly reduces the updated amplitude of parameters. Furthermore, the learned weights of the CNN-based model were updated to almost the state of convergence. After this, the learned weights only had a minor update. As a result, the training process was basically stable after 25 epochs.
5.6. The Generalization and Robustness of the CNN-Based Model
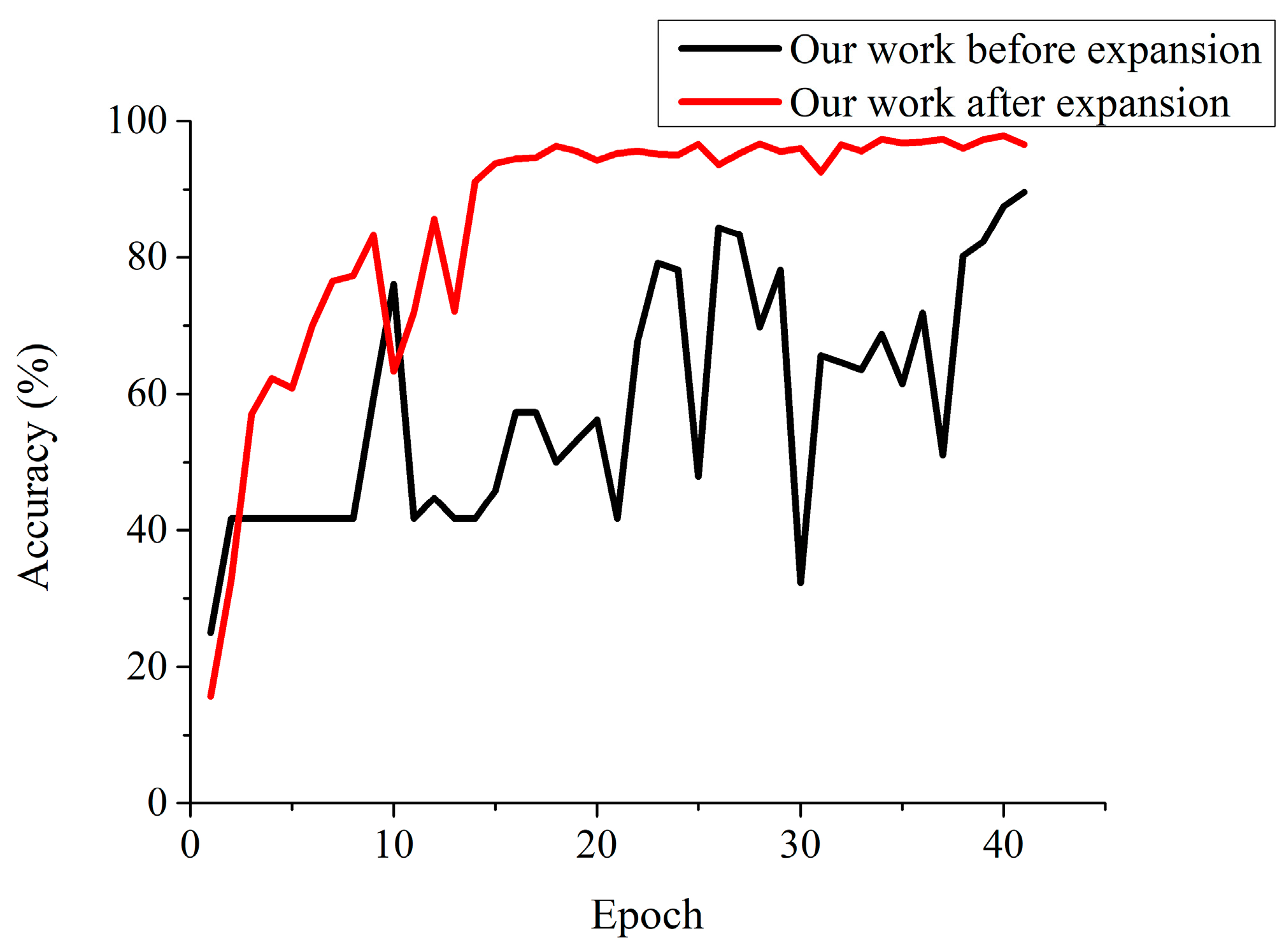
The size of the dataset has an impact on the identification accuracy of apple leaf diseases, and this paper performed two sets of experiments to estimate the effectiveness of the dataset for the proposed model, which is trained separately before and after the expansion of the dataset. From the results shown in
Figure 10, without an expanded image dataset, the proposed model has an extremely unstable training process and finally reaches a recognition rate of 86.79%. However, the proposed model with the expanded dataset achieves an accuracy of 97.62%, which improves the recognition rate by about 10.83% over that of the nonexpanded dataset.
From the known results, as shown in
Figure 10, this phenomenon is mainly due to the following reasons: (1) the expanded dataset generated by various digital image processing technologies gives the proposed CNN-based model more chances to learn appropriate layered features; (2) the diversity of images in the expanded image dataset helps to fully train the learned weights in the CNN-based model, while the smaller image dataset lacks diversity and is going to cause the overfitting problem; and (3) the preprocessing of the images simulates the real acquisition environment of the apple pathological images and, as a consequence, the CNN-based model has better identification ability for natural apple pathological images obtained from the apple orchard. The experimental result shows that the expanding dataset contributes to enhancing the generalization ability of the proposed model.
6. Conclusions
This paper has proposed a novel deep convolutional neural network model to accurately identify apple leaf diseases, which can automatically discover the discriminative features of leaf diseases and enable an end-to-end learning pipeline with high accuracy. In order to provide adequate apple pathological images, firstly, a total of 13,689 images were generated by image processing technologies, such as direction disturbance, light disturbance, and PCA jittering. Furthermore, a novel structure of a deep convolutional neural network based on the AlexNet model was designed by removing partial full connected layers, adding pooling layers, introducing the GoogLeNet Inception structure into the proposed network model, and applying the NAG algorithm to optimize network parameters to accurately identify the apple leaf diseases.
The novel deep convolutional network model was implemented in the Caffe framework on the GPU platform. Using a dataset of 13,689 images of diseased leaves, the proposed model was trained to detect apple leaf diseases. The results demonstrated are satisfactory, and the proposed model can obtain a recognition accuracy of 97.62%, which is higher than the recognition abilities of other models. Compared with the standard AlexNet model, the proposed model reduces the number of parameters greatly, has a faster convergence rate, and the accuracy of the proposed model with supplemented images is increased by 10.83% compared with the original set of diseased leaf images. The results indicated that the proposed CNN-based model can accurately identify the four common types of apple leaf diseases with high accuracy, and provides a feasible solution for identification and recognition of apple leaf diseases.
In addition, due to the restriction of biological growth laws and the current season in which the apple leaves have fallen, other diseases of apple leaves are difficult to collect. In future work, for the sake of detecting apple leaf diseases in real time, other deep neural network models, such as Faster RCNN (Regions with Convolutional Neural Network), YOLO (You Only Look Once), and SSD (Single Shot MultiBox Detector), are planned to be applied. Furthermore, more types of apple leaf diseases and thousands of high-quality natural images of apple leaf diseases still need to be gathered in the plantation in order to identify more diseases in a timely and accurate manner.
Acknowledgments
We are grateful for anonymous reviewers’ hard work and comments that allowed us to improve the quality of this paper. This work is supported by National Natural Science Foundation of China through Grant No. 61602388, by the Natural Science Basic Research Plan in Shaanxi Province of China under Grant No. 2017JM6059, by the China Postdoctoral Science Foundation under Grant No. 2017M613216, by the Postdoctoral Science Foundation of Shaanxi Province of China under Grant No. 2016BSHEDZZ121, and the Fundamental Research Funds for the Central Universities under Grants No. 2452015194 and No. 2452016081.
Author Contributions
Bin Liu contributed significantly to proposing the idea, manuscript preparation and revision, and providing the research project. Yun Zhang contributed significantly to conducting the experiment, and manuscript preparation and revision. Dongjian He and Yuxiang Li helped perform the analysis with constructive discussions.
Conflicts of Interest
We declare that we have no financial or personal relationships with other people or organizations that can inappropriately influence our work; there is no professional or other personal interest of any nature or kind in any product, service, and/or company that could be construed as influencing the position presented in, or the review of the manuscript entitled, “Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks”.
References
- Dutot, M.L.; Nelson, M.; Tyson, R.C. Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest Biol. Technol. 2013, 85, 45–56. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, G.; Li, M.Z. Management information system for apple diseases and insect pests based on GIS. Trans. Chin. Soc. Agric. Eng. 2006, 22, 150–154. [Google Scholar]
- Es-Saady, Y.; Massi, I.E.; Yassa, M.E.; Mammass, D.; Benazoun, A. Automatic recognition of plant leaves diseases based on serial combination of two SVM classifiers. In Proceedings of the 2nd International Conference on Electrical and Information Technologies, Tangiers, Morocco, 4–7 May 2016; pp. 561–566. [Google Scholar]
- Padol, P.B.; Yadav, A.A. SVM classifier based grape leaf disease detection. In Proceedings of the 2016 Advances in Signal Processing, Pune, India, 9–11 June 2016; pp. 175–179. [Google Scholar]
- Sannakki, S.S.; Rajpurohit, V.S.; Nargund, V.B.; Kumar, A.; Yallur, P.S. Diagnosis and classification of grape leaf diseases using neural networks. In Proceedings of the 4th International Conference on Computing, Tiruchengode, India, 4–6 July 2013; pp. 1–5. [Google Scholar]
- Qin, F.; Liu, D.X.; Sun, B.D.; Ruan, L.; Ma, Z.; Wang, H. Identification of alfalfa leaf diseases using image recognition technology. PLoS ONE 2016, 11, e0168274. [Google Scholar] [CrossRef] [PubMed]
- Rothe, P.R.; Kshirsagar, R.V. Cotton leaf disease identification using pattern recognition techniques. In Proceedings of the 2015 International Conference on Pervasive Computing, Pune, India, 8–10 January 2015; pp. 1–6. [Google Scholar]
- Islam, M.; Dinh, A.; Wahid, K.; Bhowmik, P. Detection of potato diseases using image segmentation and multiclass support vector machine. In Proceedings of the 30th IEEE Canadian Conference on Electrical and Computer Engineering, Windsor, ON, Canada, 30 April–3 May 2017; pp. 1–4. [Google Scholar]
- Gupta, T. Plant leaf disease analysis using image processing technique with modified SVM-CS classifier. Int. J. Eng. Manag. Technol. 2017, 5, 11–17. [Google Scholar]
- Dhakate, M.; Ingole, A.B. Diagnosis of pomegranate plant diseases using neural network. In Proceedings of the 5th National Conference on Computer Vision, Pattern Recognition, Image Processing and Graphics, Patna, India, 16–19 December 2015; pp. 1–4. [Google Scholar]
- Gavhale, M.K.R.; Gawande, U. An overview of the research on plant leaves disease detection using image processing techniques. J. Comput. Eng. 2014, 16, 10–16. [Google Scholar]
- Wang, G.; Sun, Y.; Wang, J.X. Automatic image-based plant disease severity estimation using deep learning. Comput. Intell. Neurosci. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.P.; Hughes, D.; Salathe, M. Inference of Plant Diseases from Leaf Images through Deep Learning. arXiv. 2016. Available online: https://www.semanticscholar.org/paper/Inference-of-Plant-Diseases-from-Leaf-Images-throu-Mohanty-Hughes/62163ff3cb2fbbf5361e340f042b6c288d3b8e6a (accessed on 28 December 2017).
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.M.J.; Joy, A.; Francis, J. Plant leaf disease detection using deep learning and convolutional neural network. Int. J. Eng. Sci. Comput. 2017, 7, 5324–5328. [Google Scholar]
- Lu, Y.; Yi, S.J.; Zeng, N.Y.; Liu, Y.; Zhang, Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Tan, W.X.; Zhao, C.J.; Wu, H.R. CNN intelligent early warning for apple skin lesion image acquired by infrared video sensors. High Technol. Lett. 2016, 22, 67–74. [Google Scholar]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic study of automated diagnosis of viral plant diseases using convolutional neural networks. In Proceedings of the 12th International Symposium on Visual Computing, Las Vegas, NV, USA, 12–14 December 2015; pp. 638–645. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Marcel, S. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [PubMed]
- Heisel, S.; Kovačević, T.; Briesen, H.; Schembecker, G.; Wohlgemuth, K. Variable selection and training set design for particle classification using a linear and a non-linear classifier. Chem. Eng. Sci. 2017, 173, 131–144. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.Q.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, R. Going deeper with convolutions. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 24–27 June 2014; pp. 1–9. [Google Scholar]
- Giusti, A.; Dan, C.C.; Masci, J.; Gambardella, L.M.; Schmidhuber, J. Fast image scanning with deep max-pooling convolutional neural networks. In Proceedings of the 20th IEEE International Conference on Image Processing, Melbourne, Australia, 15–18 September 2013; pp. 4034–4038. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Stochastic pooling for regularization of deep convolutional neural networks. arXiv, 2013; arXiv:1301.3557. [Google Scholar]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv. 2016. Available online: https://arxiv.org/abs/1609.04747 (accessed on 28 December 2017).
- Bahrampour, S.; Ramakrishnan, N.; Schott, L.; Shah, M. Comparative study of caffe, neon, theano, and torch for deep learning. In Proceedings of the 2016 International Conference on Learning Representations, San Juan, PR, USA, 2–5 May 2016; pp. 1–11. [Google Scholar]
- Liu, B.; He, J.R.; Geng, Y.J.; Huang, L.; Li, S. Toward emotion-aware computing: A loop selection approach based on machine learning for speculative multithreading. IEEE Access 2017, 5, 3675–3686. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).