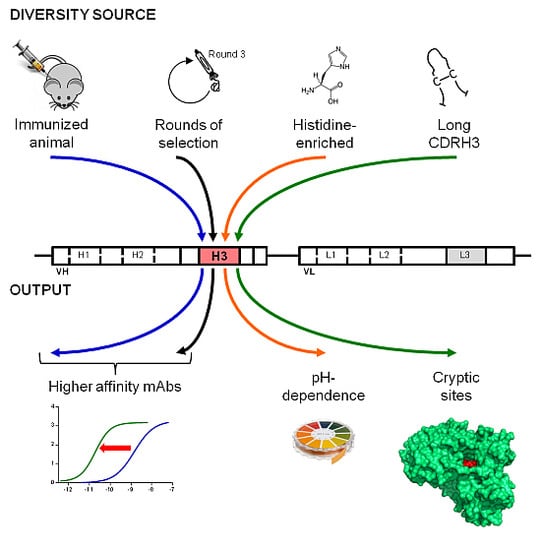
Purpose-Oriented Antibody Libraries Incorporating Tailored CDR3 Sequences
Abstract
:1. Introduction
2. Results and Discussion
2.1. Long CDRH3 Libraries for the Inhibition of Enzymes
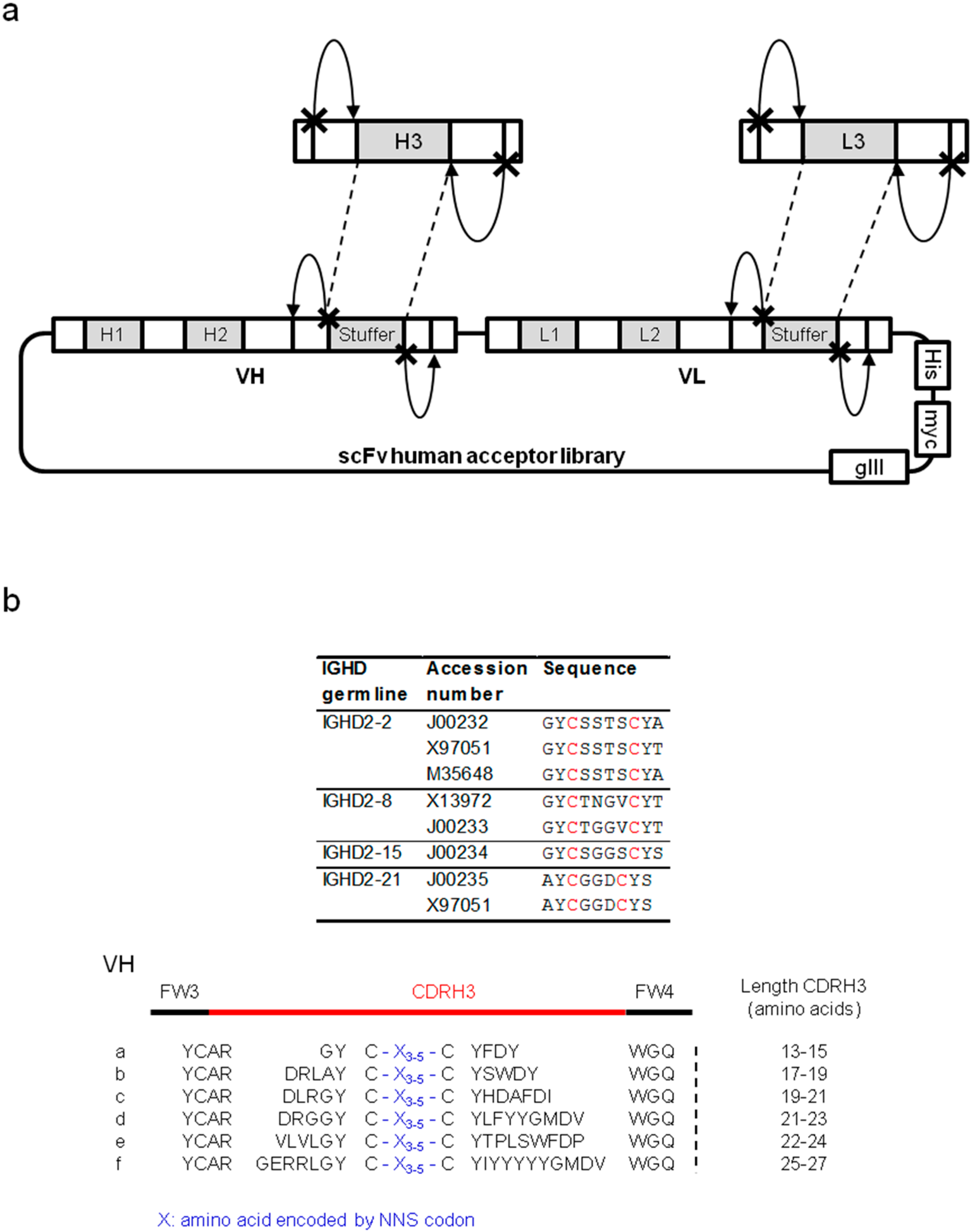
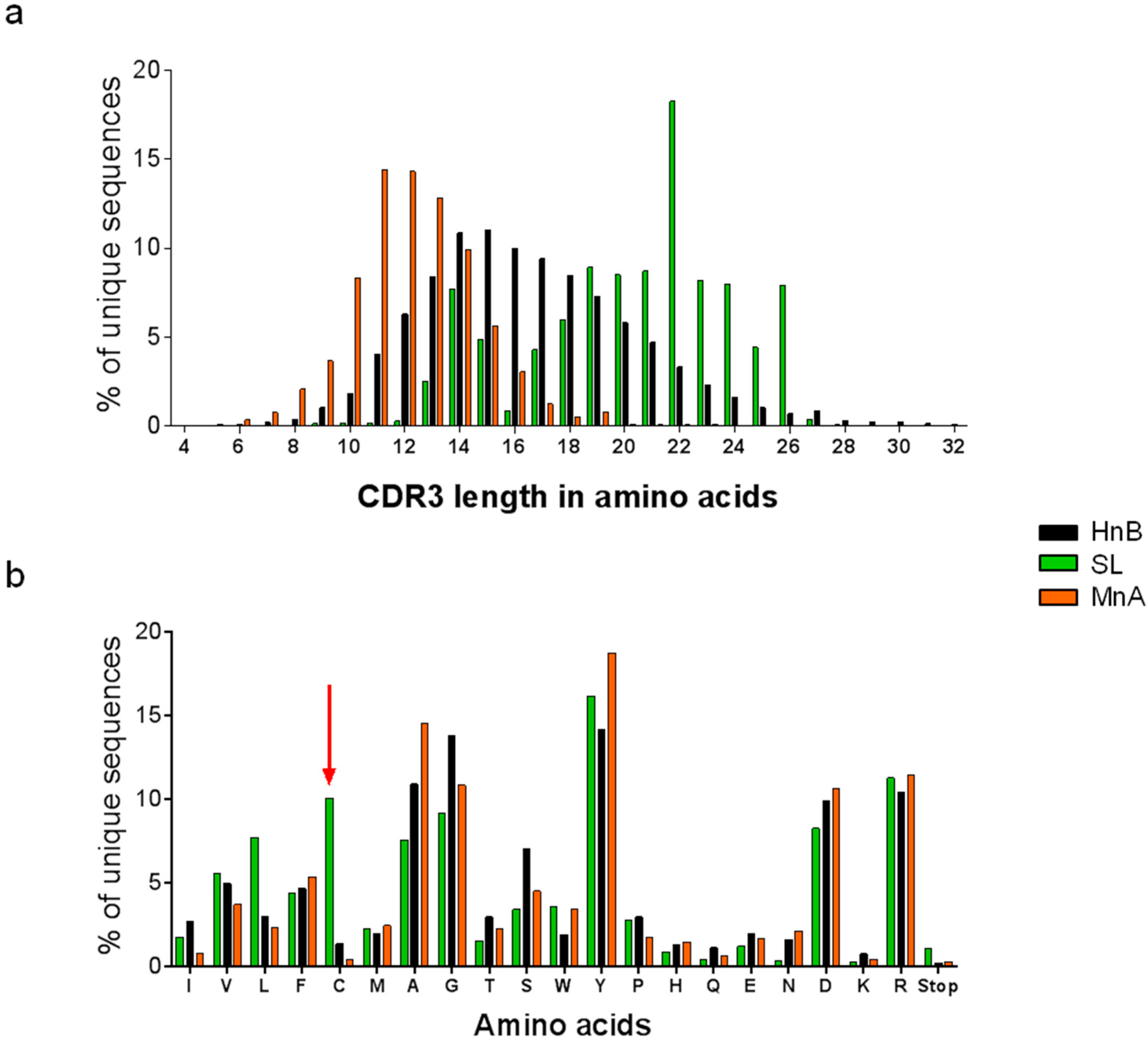
2.1.1. Characterization of Libraries by Deep Sequencing
| CDRH3 source | Library | Size | Unique CDRH3 | |
|---|---|---|---|---|
| Number | % | |||
| Human | HnB | 2.9 × 108 | 313'094 | 38.1% |
| Mouse | MnA | 2.5 × 108 | 70'993 | 13.0% |
| Synthetic long | SL | 5.2 × 109 | 398'124 | 43.6% |
| *185'453 | *23.3% | |||
2.1.2. Assessment of Library Performance by Phage Display Selections against HRP
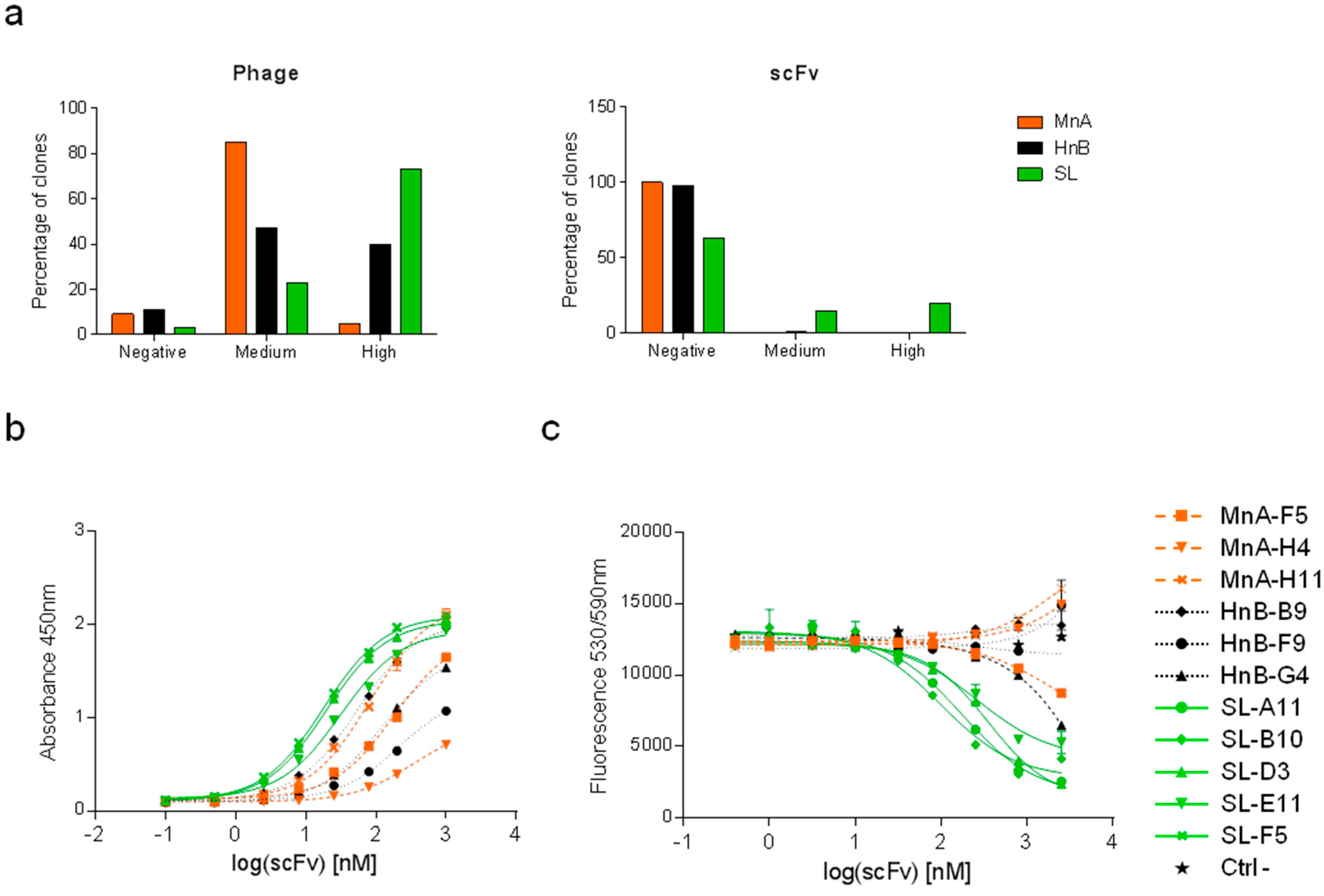
| Clone | CDRH3 | CDRL3 | CDRH3 length | HRP inhibition IC50 [nM] |
|---|---|---|---|---|
| MnA-A3 | ARDAHGWYF----------DV | GTWDMPPD-VV | 11 | - |
| MnA-F5 | ARSYANYDWF---------DY | QQSVHYRP-ST | 12 | - |
| MnA-H4 | ARLGTGTGYF---------DV | GTYDVGRV-HV | 12 | - |
| MnA-H11 | ARQGNGYY-----------AY | QQSEGVP--WT | 10 | - |
| HnB-A4 | ARPQTYYYDSSGYPDAF--DI | QQREVLP--LT | 19 | - |
| HnB-B9 | ARGWGRTAT----------DY | QQPARQWP-RT | 11 | - |
| HnB-F9 | ARDPGVLWSSSSPYYF---DY | QQDLAGWP-BT | 18 | - |
| HnB-G4 | ARGLVFDSSGYYGF-----DY | QQPSPMP--PT | 16 | 3414 |
| SL-A11 | ARDLRGYC-GLV-CYHDAFDI | GTYDGATQLPV | 19 | 165 |
| SL-B10 | ARDLRGYCSWSRLCYHDAFDI | AAYDASRV-SV | 21 | 107 |
| SL-D3 | ARDLRGYCAWSRACYHDAFDI | AAWDSSPA-VV | 21 | 412 |
| SL-E11 | ARDLRGYC-GVV-CYHDAFDI | GTYDGTERVYV | 19 | 275 |
| SL-F5 | ARDLRGYCAWSRSCYHDAFDI | AAWDSRAA-LV | 21 | 495 |
| SL-G7 | ARDLRGYC-GVF-CYHDAFDI | GTYDGSASEPV | 19 | 1948 |
2.2. Anti-Fc Antibodies with pH-Dependent Binding Properties
2.2.1. Construction of a Synthetic Library Biased Towards Histidine
2.2.2. Selections against Human Fc Fragments
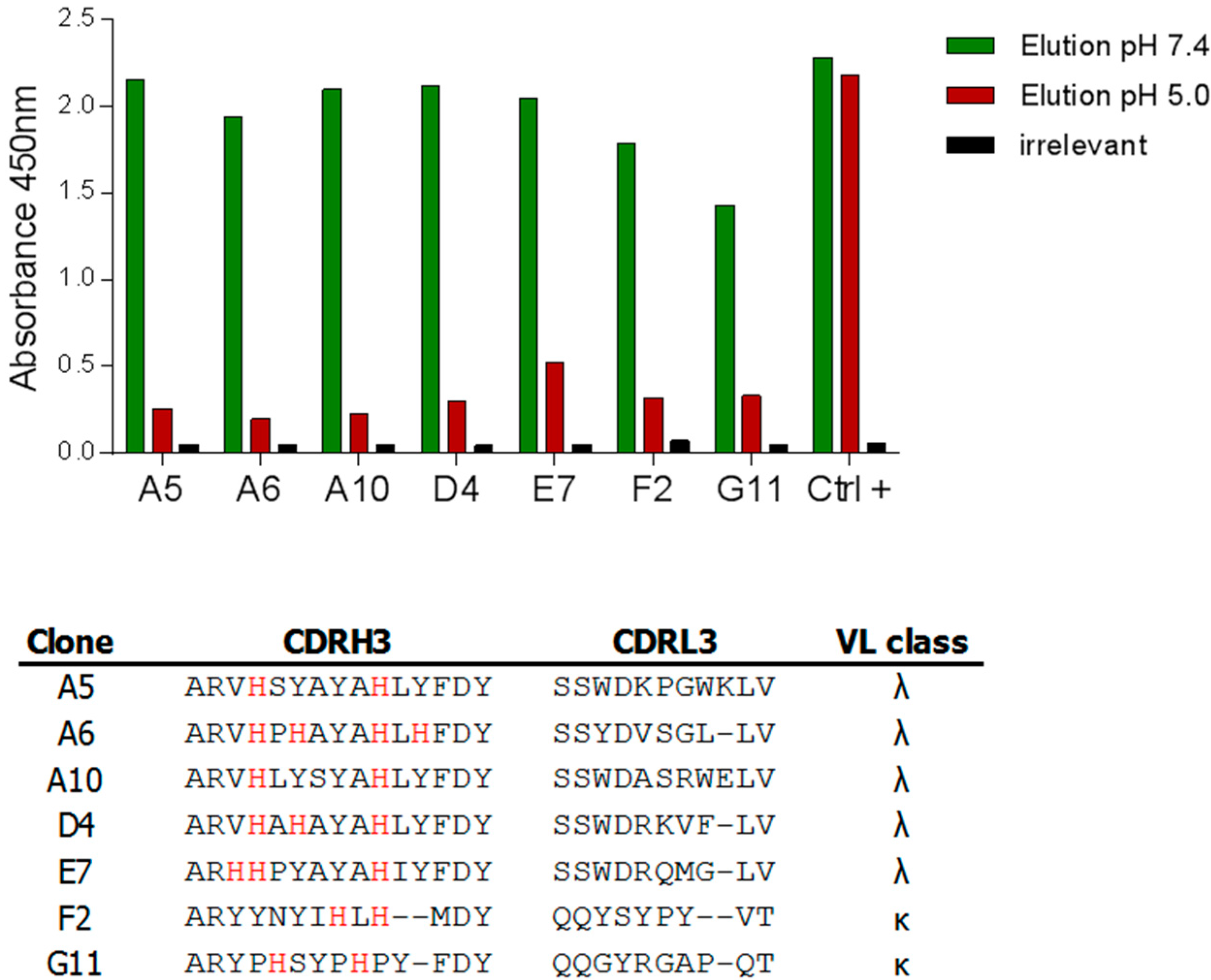
2.2.3. Characterization of pH-Dependent Anti-hFc Candidates
3. Experimental Section
3.1. Reagents
3.2. Construction of the Synthetic Long Library and Sequencing

3.3. Phage Display Selections against HRP
3.4. Screening ELISA on HRP
3.5. Dose Response Functional Assay of HRP Activity
3.6. Construction of Histidine-Enriched Libraries
3.7. pH-Dependent Phage Display Selections and Screening
3.8. Expression and Purification of Antibodies
3.9. Characterization of pH-Dependent Anti-hFc IgGs
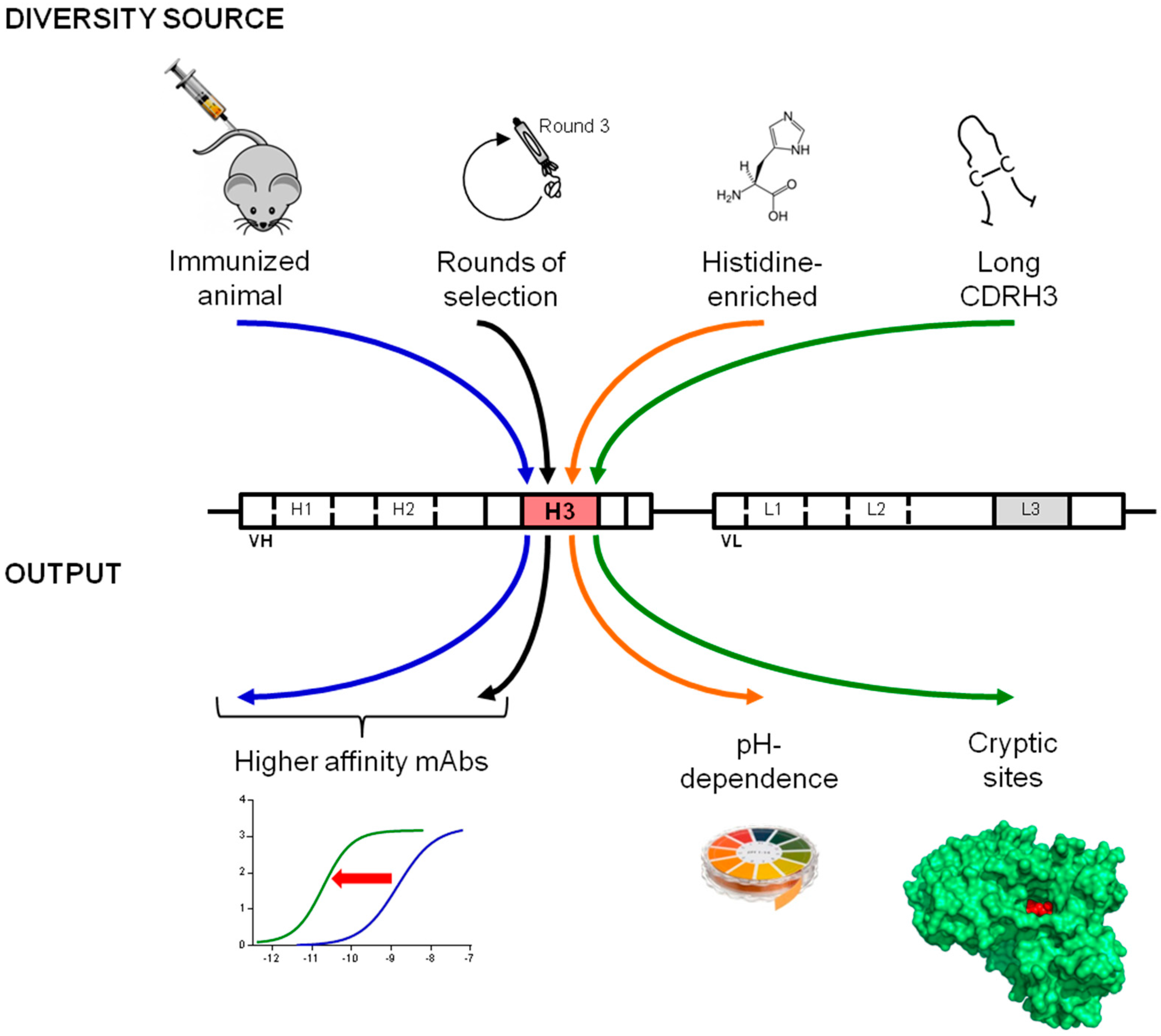
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Geyer, C.R.; McCafferty, J.; Dubel, S.; Bradbury, A.R.; Sidhu, S.S. Recombinant antibodies and in vitro selection technologies. Methods Mol. Biol. 2012, 901, 11–32. [Google Scholar] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Amstutz, P.; Pluckthun, A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 2005, 23, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, A.R.; Sidhu, S.; Dubel, S.; McCafferty, J. Beyond natural antibodies: The power of in vitro display technologies. Nat. Biotechnol. 2011, 29, 245–254. [Google Scholar] [CrossRef] [PubMed]
- de Haard, H.J.; van, N.N.; Reurs, A.; Hufton, S.E.; Roovers, R.C.; Henderikx, P.; de Bruine, A.P.; Arends, J.W.; Hoogenboom, H.R. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 1999, 274, 18218–18230. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.; Lowe, D.; Edwards, B.; Welsh, F.; Dilks, T.; Hardman, C.; Vaughan, T. Modelling the human immune response: Performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng. Des. Sel. 2009, 22, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Nissim, A.; Hoogenboom, H.R.; Tomlinson, I.M.; Flynn, G.; Midgley, C.; Lane, D.; Winter, G. Antibody fragments from a 'single pot' phage display library as immunochemical reagents. EMBO J. 1994, 13, 692–698. [Google Scholar] [PubMed]
- Prassler, J.; Thiel, S.; Pracht, C.; Polzer, A.; Peters, S.; Bauer, M.; Norenberg, S.; Stark, Y.; Kolln, J.; Popp, A.; et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J. Mol. Biol. 2011, 413, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Hust, M.; Dubel, S. Mating antibody phage display with proteomics. Trends Biotechnol. 2004, 22, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Persson, M.A.; Caothien, R.H.; Burton, D.R. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc. Natl. Acad. Sci. USA 1991, 88, 2432–2436. [Google Scholar] [CrossRef] [PubMed]
- Tikunova, N.; Dubrovskaya, V.; Morozova, V.; Yun, T.; Khlusevich, Y.; Bormotov, N.; Laman, A.; Brovko, F.; Shvalov, A.; Belanov, E. The neutralizing human recombinant antibodies to pathogenic Orthopoxviruses derived from a phage display immune library. Virus Res. 2012, 163, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Cobaugh, C.W.; Almagro, J.C.; Pogson, M.; Iverson, B.; Georgiou, G. Synthetic antibody libraries focused towards peptide ligands. J. Mol. Biol. 2008, 378, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Lantto, J.; Ohlin, M. A focused antibody library for improved hapten recognition. J. Mol. Biol. 2006, 357, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, P.M.; Neri, D.; Winter, G. Towards the design of an antibody that recognises a given protein epitope. J. Mol. Biol. 1999, 285, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Chaparro-Riggers, J.; Liang, H.; DeVay, R.M.; Bai, L.; Sutton, J.E.; Chen, W.; Geng, T.; Lindquist, K.; Casas, M.G.; Boustany, L.M.; et al. Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J. Biol. Chem. 2012, 287, 11090–11097. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Mimoto, F.; Hattori, K. pH-dependent antigen-binding antibodies as a novel therapeutic modality. Biochim. Biophys. Acta 2014, 1844, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Ravn, U.; Gueneau, F.; Baerlocher, L.; Osteras, M.; Desmurs, M.; Malinge, P.; Magistrelli, G.; Farinelli, L.; Kosco-Vilbois, M.H.; Fischer, N. By-passing in vitro screening--next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010, 38, e193. [Google Scholar] [CrossRef] [PubMed]
- Venet, S.; Ravn, U.; Buatois, V.; Gueneau, F.; Calloud, S.; Kosco-Vilbois, M.; Fischer, N. Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences. PLoS One 2012, 7, e43471. [Google Scholar] [CrossRef] [PubMed]
- Bonvin, P.; Venet, S.; Fontaine, G.; Ravn, U.; Gueneau, F.; Kosco-Vilbois, M.; Proudfoot, A.E.; Fischer, N. De novo isolation of antibodies with pH-dependent binding properties. MAbs 2015, 7, 294–302. [Google Scholar] [PubMed]
- Laskowski, R.A.; Luscombe, N.M.; Swindells, M.B.; Thornton, J.M. Protein clefts in molecular recognition and function. Protein Sci. 1996, 5, 2438–2452. [Google Scholar] [PubMed]
- Cope, P.A.; Mooser, G. Antibodies against active-site peptides common to glucosyltransferases of mutans streptococci. Infect. Immun. 1993, 61, 4814–4817. [Google Scholar] [PubMed]
- Conrath, K.E.; Lauwereys, M.; Galleni, M.; Matagne, A.; Frere, J.M.; Kinne, J.; Wyns, L.; Muyldermans, S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 2001, 45, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, A.; Transue, T.R.; Ghahroudi, M.A.; Thi, M.H.; Poortmans, F.; Hamers, R.; Muyldermans, S.; Wyns, L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 1996, 3, 803–811. [Google Scholar] [CrossRef] [PubMed]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Lauwereys, M.; Arbabi, G.M.; Desmyter, A.; Kinne, J.; Holzer, W.; De, G.E.; Wyns, L.; Muyldermans, S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998, 17, 3512–3520. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O'Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Ohshima, N.; Stanfield, R.L.; Yu, W.; Iba, Y.; Okuno, Y.; Kurosawa, Y.; Wilson, I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014, 5, 3614. [Google Scholar] [PubMed]
- Rouet, R.; Dudgeon, K.; Christie, M.; Langley, D.; Christ, D. Fully human VH single domains that rival the stability and cleft recognition of camelid antibodies. J. Biol. Chem. 2015, 290, 11905–11917. [Google Scholar] [CrossRef] [PubMed]
- Zemlin, M.; Klinger, M.; Link, J.; Zemlin, C.; Bauer, K.; Engler, J.A.; Schroeder, H.W., Jr.; Kirkham, P.M. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 2003, 334, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Berglund, G.I.; Carlsson, G.H.; Smith, A.T.; Szoke, H.; Henriksen, A.; Hajdu, J. The catalytic pathway of horseradish peroxidase at high resolution. Nature 2002, 417, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.F.; Ma, J.; Winter, C.; Bayer, R. Recovery and purification process development for monoclonal antibody production. MAbs 2010, 2, 480–499. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.P.; Giudicelli, V.; Ginestoux, C.; Bodmer, J.; Muller, W.; Bontrop, R.; Lemaitre, M.; Malik, A.; Barbie, V.; Chaume, D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999, 27, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Madden, T. The BLAST Sequence Analysis Tool. Available online: http://www.ncbi.nlm.nih.gov/books/NBK153387/ (accessed on 12 May 2015).
- Ravn, U.; Didelot, G.; Venet, S.; Ng, K.T.; Gueneau, F.; Rousseau, F.; Calloud, S.; Kosco-Vilbois, M.; Fischer, N. Deep sequencing of phage display libraries to support antibody discovery. Methods 2013, 60, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Elson, G.; Magistrelli, G.; Dheilly, E.; Fouque, N.; Laurendon, A.; Gueneau, F.; Ravn, U.; Depoisier, J.F.; Moine, V.; et al. Exploiting light chains for the scalable generation and platform purification of native human bispecific IgG. Nat. Commun. 2015, 6, 6113. [Google Scholar] [CrossRef] [PubMed]
- Deruaz, M.; Bonvin, P.; Severin, I.C.; Johnson, Z.; Krohn, S.; Power, C.A.; Proudfoot, A.E. Evasin-4, a tick-derived chemokine-binding protein with broad selectivity can be modified for use in preclinical disease models. FEBS J. 2013, 280, 4876–4887. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ekiert, D.C.; Ahmad, I.; Yu, W.; Zhang, Y.; Bazirgan, O.; Torkamani, A.; Raudsepp, T.; Mwangi, W.; Criscitiello, M.F.; et al. Reshaping antibody diversity. Cell 2013, 153, 1379–1393. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonvin, P.; Venet, S.; Kosco-Vilbois, M.; Fischer, N. Purpose-Oriented Antibody Libraries Incorporating Tailored CDR3 Sequences. Antibodies 2015, 4, 103-122. https://doi.org/10.3390/antib4020103
Bonvin P, Venet S, Kosco-Vilbois M, Fischer N. Purpose-Oriented Antibody Libraries Incorporating Tailored CDR3 Sequences. Antibodies. 2015; 4(2):103-122. https://doi.org/10.3390/antib4020103
Chicago/Turabian StyleBonvin, Pauline, Sophie Venet, Marie Kosco-Vilbois, and Nicolas Fischer. 2015. "Purpose-Oriented Antibody Libraries Incorporating Tailored CDR3 Sequences" Antibodies 4, no. 2: 103-122. https://doi.org/10.3390/antib4020103
APA StyleBonvin, P., Venet, S., Kosco-Vilbois, M., & Fischer, N. (2015). Purpose-Oriented Antibody Libraries Incorporating Tailored CDR3 Sequences. Antibodies, 4(2), 103-122. https://doi.org/10.3390/antib4020103