snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management
Abstract
:1. Introduction
2. snoRNAs: An Age-Old Class of Non-Coding RNAs Recently Rediscovered
2.1. Abundance and Diversity of Human snoRNAs
2.2. snoRNAs Are More Than Just Major Actors of Ribosome Biogenesis
2.3. Clinical Significance of snoRNAs in Human Diseases
3. Lung Cancer: A Long Quest to Overcome a Major Killer
3.1. A Clinical Overview of Lung Cancer
3.2. Histological and Genomic Classification of Lung Cancer
3.3. Therapies and Limitations
4. Altered Expression of snoRNAs in Lung Cancer
4.1. snoRNA Profiling in Normal and Tumoral Lung Tissues
4.2. Circulating snoRNA Profiling in Healthy and Lung Cancer Patients
4.3. Clinical Significance of Altered snoRNAs in Lung Cancer
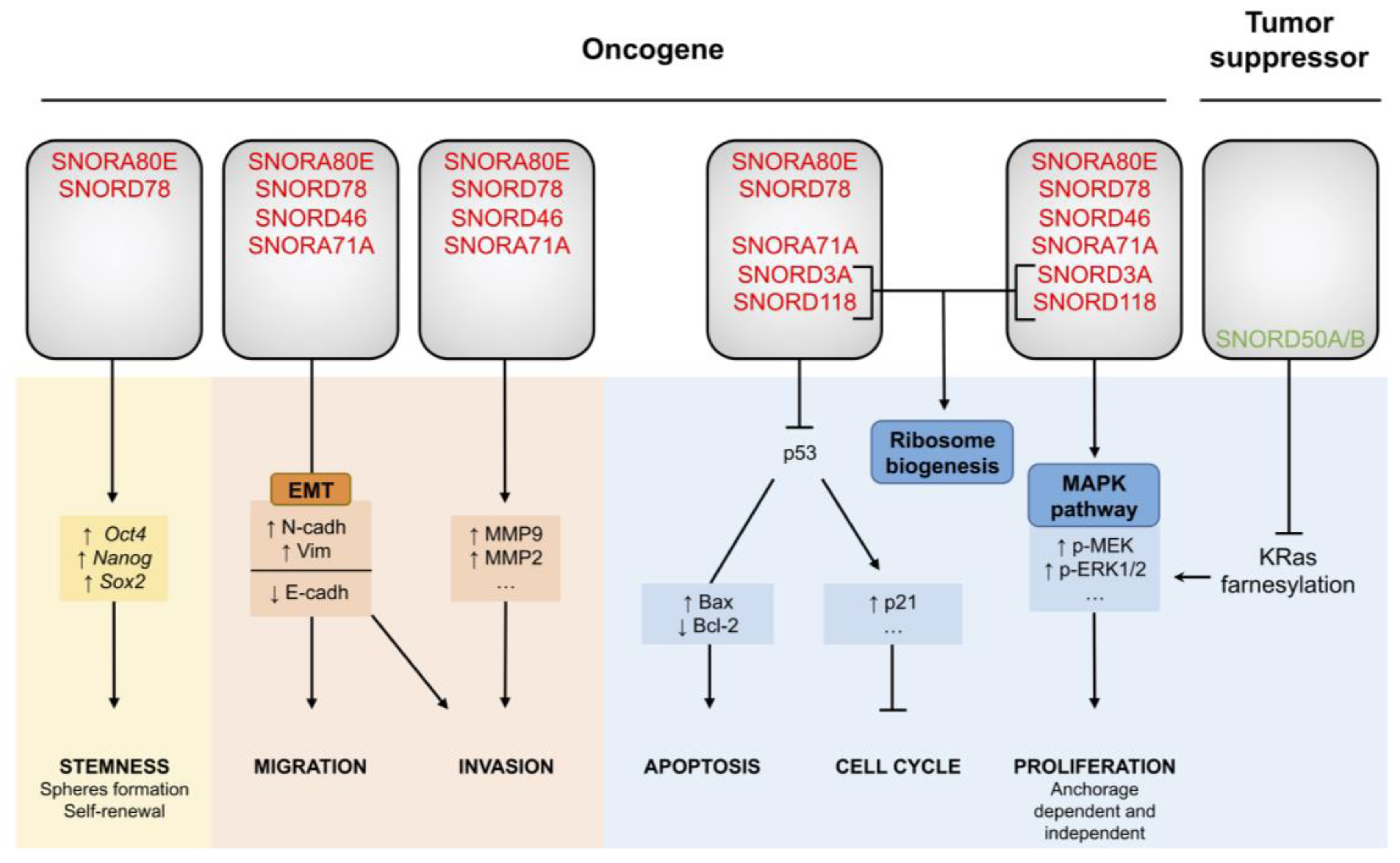
5. snoRNA Dysregulation Contributes to Lung Cancer Tumorigenesis
5.1. SNORA80E Inhibits Apoptosis and Supports Stemness
5.2. SNORD78 Triggers Cancer Cell Plasticity
5.3. SNORA71A: A Regulator of MAPK Signaling
5.4. SNORD50A/B, Inhibitors of the Normal KRas Pathway
5.5. SNORD3A and SNORD118: From Ribosome Biogenesis to Tumorigenesis
6. The Future of snoRNAs in the Field of Lung Cancer
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Tar. 2016, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, J.; Li, Y.; Liu, C.-J.; Guo, A.-Y.; Lin, C.; Correspondence, H. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, S.B.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7. [Google Scholar]
- Neel, D.S.; Bivona, T.G. Resistance is futile: Overcoming resistance to targeted therapies in lung adenocarcinoma. npj Precis. Oncol. 2017, 1. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, E.; Fournier, M. The Small Nucleolar RNAs. Annu. Rev. Biochem. 1995, 64, 897–934. [Google Scholar] [CrossRef]
- Stern, H.; Johnston, F.; Setterfield, G. Some chemical properties of isolated pea nucleoli. J. Biophys. Biochem. Cytol. 1959, 6, 57–60. [Google Scholar] [CrossRef]
- Kufel, J.; Grzechnik, P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [Green Version]
- Lafontaine, D.L.J. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef]
- Jorjani, H.; Kehr, S.; Jedlinski, D.J.; Gumienny, R.; Hertel, J.; Stadler, P.F.; Zavolan, M.; Gruber, A.R. An updated human snoRNAome. Nucleic Acids Res. 2016, 44, 5068–5082. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakin, A.G.; Smith, L. The RNA World of the Nucleolus: Two Major Families of Small RNAs Defined by Different Box Elements with Related Functions. Cell 1996, 86, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, E.; Fournier, M.J. The snoRNPs and Related Machines: Ancient Devices That Mediate Maturation of rRNA and Other RNAs. In Madame Curie Bioscience Database; Olson, M.O.J., Ed.; Landes Bioscience: Austin, TX, USA, 2011; pp. 223–258. [Google Scholar]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Monaco, P.; Marcel, V.; Diaz, J.J.; Catez, F. 2′-O-methylation of ribosomal RNA: Towards an epitranscriptomic control of translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef] [Green Version]
- Tollervey, D.; Kiss, T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997, 9, 337–342. [Google Scholar] [CrossRef]
- Therizols, G.; Laforêts, F.; Marcel, V.; Catez, F.; Bouvet, P.; Diaz, J.J. Ribosomal RNA Methylation and Cancer. In Epigenetic Cancer Therapy; Academic Press: Cambridge, MA, USA, 2015; pp. 115–139. ISBN 9780128002247. [Google Scholar]
- Kass, S.; Tyc, K.; Steitz, J.A.; Sollner-Webb, B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 1990, 60, 897–908. [Google Scholar] [CrossRef]
- Langhendries, J.-L.; Nicolas, E.; Doumont, G.; Goldman, S.; Lafontaine, D.L.J. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget 2016, 7, 59519–59534. [Google Scholar] [CrossRef] [Green Version]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef] [Green Version]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human snoRNA with MicroRNA-Like Functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Huang, C.; Shi, J.; Guo, Y.; Huang, W.; Huang, S.; Ming, S.; Wu, X.; Zhang, R.; Ding, J.; Zhao, W.; et al. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017, 45, 8647–8660. [Google Scholar] [CrossRef] [Green Version]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, T.; Del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.; Cheung, S.W.; Beaudet, A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008, 40, 719–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. BioEssays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Köhn, M.; Misiak, D.; Bäumer, N.; Cui, C.; Göllner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.M.; Thompson, I.; Yuan, Z.M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef]
- Davanian, H.; Balasiddaiah, A.; Heymann, R.; Sundström, M.; Redenström, P.; Silfverberg, M.; Brodin, D.; Sällberg, M.; Lindskog, S.; Kruger Weiner, C.; et al. Ameloblastoma RNA profiling uncovers a distinct non-coding RNA signature. Oncotarget 2017, 8, 4530–4542. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, M.; Yan, Y.; Kuang, Y.; Li, P.; Zheng, W.; Liu, H.; Jia, B. Identification of Eight Small Nucleolar RNAs as Survival Biomarkers and Their Clinical Significance in Gastric Cancer. Front. Oncol. 2019, 9, 788–794. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef] [Green Version]
- Mannoor, K.; Liao, J.; Jiang, F. Small nucleolar RNAs in cancer. Biochim. Biophys. 2012, 1826, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Thorenoor, N.; Slaby, O. Small nucleolar RNAs functioning and potential roles in cancer. Tumor Biol. 2014, 36, 41–53. [Google Scholar] [CrossRef]
- Lung Cancer Prevention (PDQ®) - Patient Version. Available online: https://www.cancer.gov/types/lung/patient/lung-prevention-pdq#section/all (accessed on 13 February 2020).
- Barta, J.A.; Henschke, C.I.; Flores, R.M.; Yip, R.; Yankelevitz, D.F.; Powell, C.A. Lung Cancer Diagnosis by Fine Needle Aspiration Is Associated With Reduction in Resection of Nonmalignant Lung Nodules. Ann. Thorac. Surg. 2017, 103, 1795–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera, M.P.; Mehta, A.C.; Wahidi, M.M. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest 2013, 143, e142S–e165S. [Google Scholar] [CrossRef] [PubMed]
- Mlika, M.; Hofman, P.; Dziri, C.; Mezni, F. Liquid biopsy in lung cancer. Tunis. Med. 2017, 95, 965–971. [Google Scholar] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.M.; Austin, J.H.; Beth Beasley, M.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [Green Version]
- Gurda, G.T.; Zhang, L.; Wang, Y.; Chen, L.; Geddes, S.; Cho, W.C.; Askin, F.; Gabrielson, E.; Li, Q.K. Utility of five commonly used immunohistochemical markers TTF-1, Napsin A, CK7, CK5/6 and P63 in primary and metastatic adenocarcinoma and squamous cell carcinoma of the lung: A retrospective study of 246 fine needle aspiration cases. Clin. Transl. Med. 2015, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Kashima, J.; Kitadai, R.; Okuma, Y. Molecular and morphological profiling of lung cancer: A foundation for “next-generation” pathologists and oncologists. Cancers 2019, 11, 599. [Google Scholar] [CrossRef] [Green Version]
- Balsara, B.R.; Testa, J.R. Chromosomal imbalances in human lung cancer. Oncogene 2002, 21, 6877–6883. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.J.; Shaw, A.T. Resisting Resistance: Targeted Therapies in Lung Cancer. Trends Cancer 2016, 2, 350–364. [Google Scholar] [CrossRef] [Green Version]
- Shames, D.S.; Wistuba, I.I. The evolving genomic classification of lung cancer. J. Pathol. 2014, 232, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Schallenberg, S.; Merkelbach-Bruse, S.; Buettner, R. Lung cancer as a paradigm for precision oncology in solid tumours. Virchows Arch. 2017, 471, 221–233. [Google Scholar] [CrossRef]
- Gallant, J.N.; Lovly, C.M. Established, emerging and elusive molecular targets in the treatment of lung cancer. J. Pathol. 2018, 244, 565–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, A.N.; Niederst, M.J.; Archibald, H.L.; Gomez-Caraballo, M.; Siddiqui, F.M.; Mulvey, H.E.; Maruvka, Y.E.; Ji, F.; Bhang, H.E.C.; Radhakrishna, V.K.; et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 2016, 22, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Sanmamed, M.F.; Hastings, K.; Politi, K.; Rimm, D.L.; Chen, L.; Melero, I.; Schalper, K.A.; Herbst, R.S. Immunotherapy in non-small cell lung cancer: Facts and hopes. Clin. Cancer Res. 2019, 25, 4592–4602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, T.S.K.; Wu, Y.L.; Kudaba, I.; Kowalski, D.M.; Cho, B.C.; Turna, H.Z.; Castro, G.; Srimuninnimit, V.; Laktionov, K.K.; Bondarenko, I.; et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial. Lancet 2019, 393, 1819–1830. [Google Scholar] [CrossRef]
- Liao, J.; Yu, L.; Mei, Y.; Guarnera, M.; Shen, J.; Li, R.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 2010, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Ma, J.; Mannoor, K.; Guarnera, M.A.; Shetty, A.; Zhan, M.; Xing, L.; Stass, S.A.; Jiang, F. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int. J. Cancer 2015, 136, E623–E629. [Google Scholar] [CrossRef] [PubMed]
- Nogueira Jorge, N.A.; Wajnberg, G.; Ferreira, C.G.; De Sa Carvalho, B.; Passetti, F. snoRNA and piRNA expression levels modified by tobacco use in women with lung adenocarcinoma. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Chen, L.; Feng, K.-Y.; Hu, X.-H.; Zhang, Y.-H.; Kong, X.-Y.; Huang, T.; Cai, Y.-D. Analysis of Expression Pattern of snoRNAs in Different Cancer Types with Machine Learning Algorithms. Int. J. Mol. Sci. 2019, 20, 2185. [Google Scholar] [CrossRef] [Green Version]
- Templeton, A.K.; Miyamoto, S.; Babu, A.; Munshi, A.; Ramesh, R. Cancer stem cells: Progress and challenges in lung cancer. Stem Cell Investig. 2014, 1. [Google Scholar]
- Vallette, F.; Olivier, C.; Lezot, F.; Oliver, L. Dormant, quiescent, tolerant and persister cells: Four synonyms for the same target in cancer. Biochem. Pharmacol. 2019, 162, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Dawson, C.C.; Intapa, C.; Jabra-Rizk, M.A. “Persisters”: Survival at the cellular level. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raha, D.; Wilson, T.R.; Peng, J.; Peterson, D.; Yue, P.; Evangelista, M.; Wilson, C.; Merchant, M.; Settleman, J. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 2014, 74, 3579–3590. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Qi, Q.; Khanna, A.; Todd, N.W.; Deepak, J.; Lingxiao, X.; Huijun, W.; Zhenqiu, L.; Yun, S.; Stass, S.A.; et al. Aldehyde dehydrogenase 1 is a tumor stem cell-Associated marker in lung cancer. Mol. Cancer Res. 2009, 7, 330–338. [Google Scholar]
- Mannoor, K.; Shen, J.; Liao, J.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol. Cancer 2014, 13. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Liao, J.; Gao, L.; Shen, J.; Guarnera, M.A.; Zhan, M.; Fang, H.B.; Stass, S.A.; Jiang, F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget 2016, 7, 5131–5142. [Google Scholar] [CrossRef] [Green Version]
- Sadik, N.; Cruz, L.; Gurtner, A.; Rodosthenous, R.S.; Dusoswa, S.A.; Ziegler, O.; Van Solinge, T.S.; Wei, Z.; Salvador-Garicano, A.M.; Gyorgy, B.; et al. Extracellular RNAs: A new awareness of old perspectives. Methods Mol. Biol. 2018, 1740, 1–15. [Google Scholar]
- Shen, J.; Todd, N.W.; Zhang, H.; Yu, L.; Lingxiao, X.; Mei, Y.; Guarnera, M.; Liao, J.; Chou, A.; Lu, C.L.; et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab. Investig. 2011, 91, 579–587. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Todd, N.W.; Liu, Z.; Zhan, M.; Fang, H.B.; Peng, H.; Alattar, M.; Deepak, J.; Stass, S.A.; Jiang, F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 2010, 67, 170–176. [Google Scholar] [CrossRef] [Green Version]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelyeva, A.V.; Kuligina, E.V.; Bariakin, D.N.; Kozlov, V.V.; Ryabchikova, E.I.; Richter, V.A.; Semenov, D.V. Variety of RNAs in Peripheral Blood Cells, Plasma, and Plasma Fractions. Biomed. Res. Int. 2017, 2017, 7404912. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Guarnera, M.A.; Fang, H.B.; Jiang, F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol. Cancer 2016, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, G.; Zeng, Z.; Sun, W.; Li, S.; You, C.; Tang, F.; Peng, S.; Ma, S.; Luo, Y.; Xu, J.; et al. Small nucleolar RNA 71A promotes lung cancer cell proliferation, migration and invasion via MAPK/ERK pathway. J. Cancer 2019, 10, 2261–2275. [Google Scholar] [CrossRef] [Green Version]
- Siprashvili, Z.; Webster, D.E.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat. Genet. 2016, 48, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Okugawa, Y.; Toiyama, Y.; Toden, S.; Mitoma, H.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut 2017, 66, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar] [CrossRef]
- Yi, C.; Wan, X.; Zhang, Y.; Fu, F.; Zhao, C.; Qin, R.; Wu, H.; Li, Y.; Huang, Y. SNORA42 enhances prostate cancer cell viability, migration and EMT and is correlated with prostate cancer poor prognosis. Int. J. Biochem. Cell Biol. 2018, 102, 138–150. [Google Scholar] [CrossRef]
- Zheng, D.; Zhang, J.; Ni, J.; Luo, J.; Wang, J.; Tang, L.; Zhang, L.; Wang, L.; Xu, J.; Su, B.; et al. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2015, 34. [Google Scholar] [CrossRef] [Green Version]
- Sahai, E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 2005, 15, 87–96. [Google Scholar] [CrossRef]
- Ma, C.; Shi, X.; Zhu, Q.; Li, Q.; Liu, Y.; Yao, Y.; Song, Y. The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumor Biol. 2016, 37, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Aerts, J.; Den Hamer, B.; Van IJcken, W.; Den Bakker, M.; Riegman, P.; Van der Leest, C.; Van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The nucleolus: An emerging target for cancer therapy. Trends Mol. Med. 2013, 19, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Catez, F.; Diaz, J.J. Ribosome heterogeneity in tumorigenesis: The rRNA point of view. Mol. Cell. Oncol. 2015, 2. [Google Scholar] [CrossRef] [Green Version]
- Jantsch, M.F.; Quattrone, A.; O’Connell, M.; Helm, M.; Frye, M.; Macias-Gonzales, M.; Ohman, M.; Ameres, S.; Willems, L.; Fuks, F.; et al. Positioning Europe for the EPITRANSCRIPTOMICS challenge. RNA Biol. 2018, 15, 829–831. [Google Scholar] [CrossRef]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef] [Green Version]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. P53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and rRNA Methylation in Cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Gachet, S.; El-Chaar, T.; Avran, D.; Genesca, E.; Catez, F.; Quentin, S.; Delord, M.; Thérizols, G.; Briot, D.; Meunier, G.; et al. Deletion 6q Drives T-cell Leukemia Progression by Ribosome Modulation. Cancer Discov. 2018, 8, 1614–1631. [Google Scholar] [CrossRef] [Green Version]
- Justilien, V.; Ali, S.A.; Jamieson, L.; Yin, N.; Cox, A.D.; Der, C.J.; Murray, N.R.; Fields, A.P. Ect2-Dependent rRNA Synthesis Is Required for KRAS-TRP53-Driven Lung Adenocarcinoma. Cancer Cell 2017, 31, 256–269. [Google Scholar] [CrossRef] [Green Version]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christopher, A.; Kaur, R.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68. [Google Scholar] [PubMed]
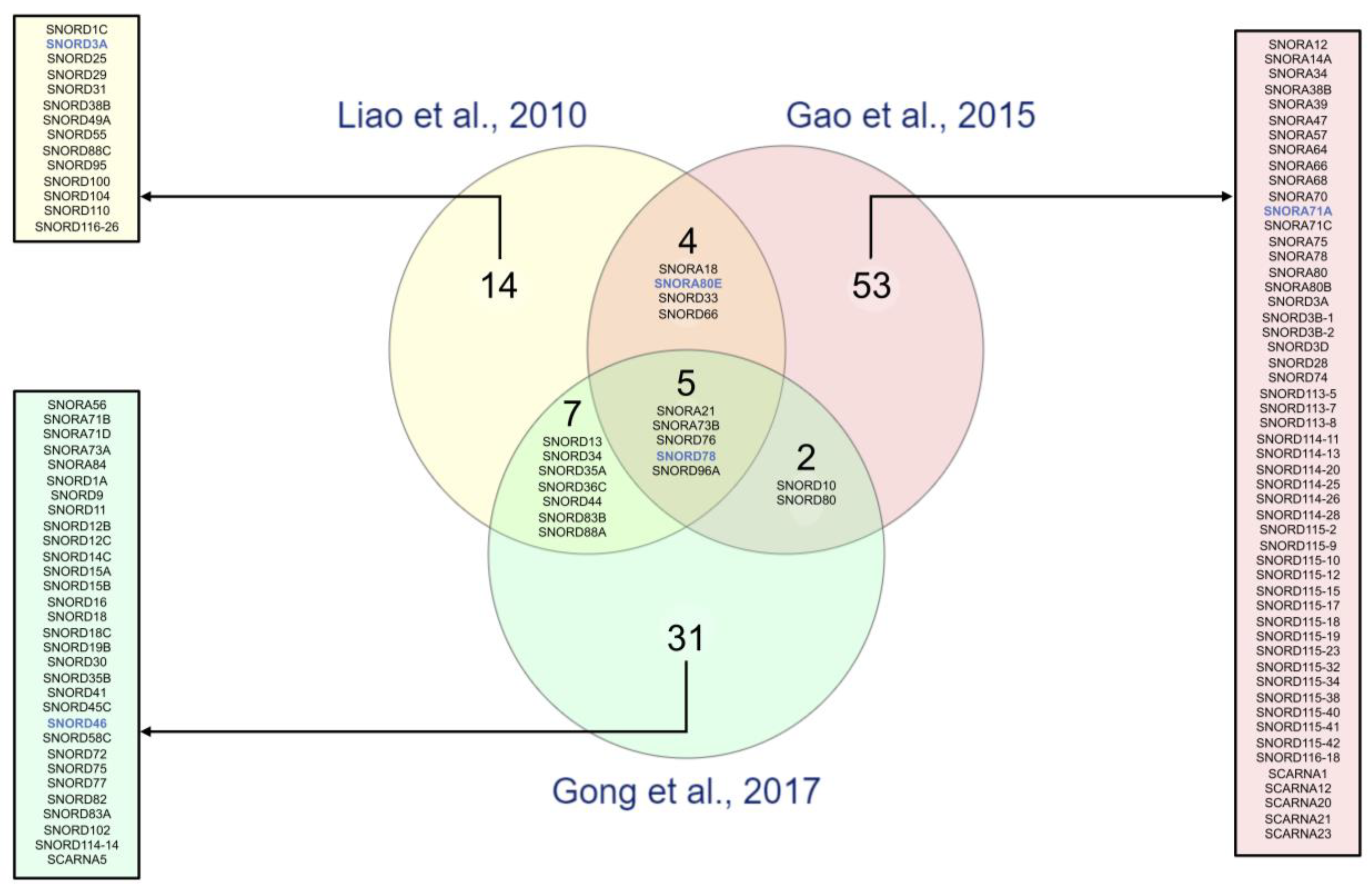
| References | Sample Type | Cohort Characteristics | Cancer Sub-Type (N) | Stage (N) | Total snoRNAs Analyzed | Technology | Fold Change Threshold | p-Value Threshold | Number of snoRNAs in Signatures | snoRNA Signature |
|---|---|---|---|---|---|---|---|---|---|---|
| a. snoRNA signatures in normal vs tumors samples | ||||||||||
| Liao et al. 2010 [48] F. Jiang lab | Lung Tissues | 22 NSCLC patient samples/22 matched normal patient samples | LUSC (11) LUAD (11) | Stage I (22) | 352 | GeneChipR Arrays (Affymetrix) | ≥|1.0-fold change| | p < 0.01 | 31 | SNORA18, SNORA21, SNORA80E, SNORA73B, SNORD1C, RNU3P2, SNORD13, SNORD25, SNORD29, SNORD31, SNORD33, SNORD34, SNORD35A, SNORD36C, SNORD38B (1), SNORD44, SNORD49A, SNORD55, SNORD66, SNORD76, SNORD78, SNORD83B, SNORD88A, SNORD88C, SNORD95, SNORD96A, SNORD100, SNORD104, SNORD110, SNORD116-26 |
| ≥|1.5-fold change| | p < 0.01 | 6 * | SNORA80E, SNORA73B, SNORD33, SNORD66, SNORD76, SNORD78 | |||||||
| Plasma | 37 NSCLC patient samples/37 non-cancerous samples (22 healthy donors and 26 COPD) | LUSC (16) LUAD (21) | Stage I (10) Stage II (12) Stage III-IV (15) | 6 * | RT-qPCR | NA | p < 0.01 | 3 | SNORD33, SNORD66, SNORD76 | |
| Gao et al. 2015 [49] F. Jiang lab | Lung Tissues | 12 NSCLC patient samples/12 matched normal patient samples | LUSC (6) LUAD (6) | Stage I (12) | 458 | Deep sequencing (Illumina® Genome Analyzer IIx) | ≥|2.0-fold change| | p ≤ 0.001 | 68 | SNORA12, SNORA14A, SNORA18, SNORA21, SNORA34, SNORA38B, SNORA39, SNORA80E, SNORA47, SNORA57, SNORA64, SNORA66, SNORA68, SNORA70 (2), SNORA71A, SNORA71C, SNORA73B, SNORA75, SNORA78, SNORA80, SNORA80B, SNORD3A, SNORD3B-1, SNORD3B-2, SNORD3D, SNORD10, SNORD28, SNORD33, SNORD66, SNORD74 (1), SNORD76, SNORD78, SNORD80, SNORD96A, SNORD113-5, SNORD113-7, SNORD113-8, SNORD114-11, SNORD114-13, SNORD114-20, SNORD114-25, SNORD114-26, SNORD114-28, SNORD115-2, SNORD115-9, SNORD115-10, SNORD115-12, SNORD115-15, SNORD115-17, SNORD115-18, SNORD115-19, SNORD115-23, SNORD115-32, SNORD115-34, SNORD115-38, SNORD115-40, SNORD115-41, SNORD115-42, SNORD116-18, SCARNA1, SCARNA12, SCARNA20 (1), SCARNA21, SCARNA23 |
| ≥|3.0-fold change| | p ≤ 0.001 | 29 | SNORA12, SNORA14A, SNORA21, SNORA34, SNORA38B, SNORA39, SNORA47, SNORA64, SNORA66, SNORA68, SNORA70, SNORA71A, SNORA71C, SNORA75, SNORA78, SNORA80, SNORA80B, SNORD10, SNORD28, SNORD66, SNORD74, SNORD80, SNORD96A, SNORD113-7, SNORD113-8, SNORD114-20, SNORD114-28, SNORD115-32, SNORD115-41 | |||||||
| Su et al. 2015 [60] F. Jiang lab | Sputum | 59 NSCLC patient samples/ 61 cancer-free smoker samples | LUSC (28) LUAD (31) | Stage I (29) Stage II (30) | 6 * | RT-qPCR | NA | p < 0.05 | 4 | SNORA80E, SNORD33, SNORD66, SNORD78 |
| Gong et al. 2017 [3] L. Han lab | Lung Tissues | 91 NSCLC patient samples/91 matched normal patient samples | LUSC (45) LUAD (46) | NA | 465 | miRNA-seq (TCGA data set) | ≥|2.0-fold change| | p < 0.05 | 46 | SNORA21, SNORA56, SNORA71B, SNORA71D, SNORA73A, SNORA73B, SNORA84 (3), SNORD1A, SNORD9, SNORD10, SNORD11, SNORD12B, SNORD12C, snoU13, SNORD13, SNORD14C, SNORD15A, SNORD15B, SNORD16 (3), SNORD18, SNORD18C, SNORD19B, SNORD30, SNORD34, SNORD35A, SNORD35B, SNORD36C, SNORD41, SNORD44, SNORD45C, SNORD46, SNORD58C, SNORD72, SNORD75, SNORD76, SNORD77, SNORD78, SNORD80, SNORD82, SNORD83A, SNORD83B, SNORD88A, SNORD96A, SNORD102, SNORD114-14 (4), SCARNA5 |
| b. CSC-snoRNA signatures in patients | ||||||||||
| Mannoor et al. 2014 [59] F. Jiang lab | Lung Tissues | ALDH+ cells (TICs) sorted from 22 NSCLC tumor tissues/ALDH− cells sorted from matched 22 NSCLC tumor tissues | NA | NA | 352 | GeneChipR Arrays (Affymetrix) | ≥|3.0-fold change| | p < 0.01 | 22 | SNORA3, SNORA18, SNORA80E, SNORA61, SNORA62, SNORD1C, SNORD14E, SNORD33, SNORD34, SNORD36C, SNORD38B, SNORD44, SNORD55, SNORD66, SNORD73B, SNORD76, SNORD78, SNORD83B, SNORD88A, SNORD96A, SNORD110, SNORD116-26 |
| c. Specific snoRNAs in lung compare to other cancer types | ||||||||||
| Pan et al. 2019 [51] Y-D. Cai lab | Lung Tissues | LUSC (521) LUAD (559) | NA | 459 | miRNA-seq (TCGA data set and Gong et al. 2019 [3] data) | NA | 10 (5) | SNORA31 (ACA31) (3), SNORA47 (HBI-115) (3), SNORD7 (mgU6-47) (4), SNORD81 (U81) (4), SNORD83B (U83B) (3), SNORD99 (HBII-420) (4), SNORD115 (4), SNORD115-32 (HBII-52-32) (4), SNORD123 (3)(4) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mourksi, N.-E.-H.; Morin, C.; Fenouil, T.; Diaz, J.-J.; Marcel, V. snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management. Cells 2020, 9, 541. https://doi.org/10.3390/cells9030541
Mourksi N-E-H, Morin C, Fenouil T, Diaz J-J, Marcel V. snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management. Cells. 2020; 9(3):541. https://doi.org/10.3390/cells9030541
Chicago/Turabian StyleMourksi, Nour-El-Houda, Chloé Morin, Tanguy Fenouil, Jean-Jacques Diaz, and Virginie Marcel. 2020. "snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management" Cells 9, no. 3: 541. https://doi.org/10.3390/cells9030541






