Connecting the Dots in the Neuroglobin-Protein Interaction Network of an Unstressed and Ferroptotic Cell Death Neuroblastoma Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Stable Cell Line Generation
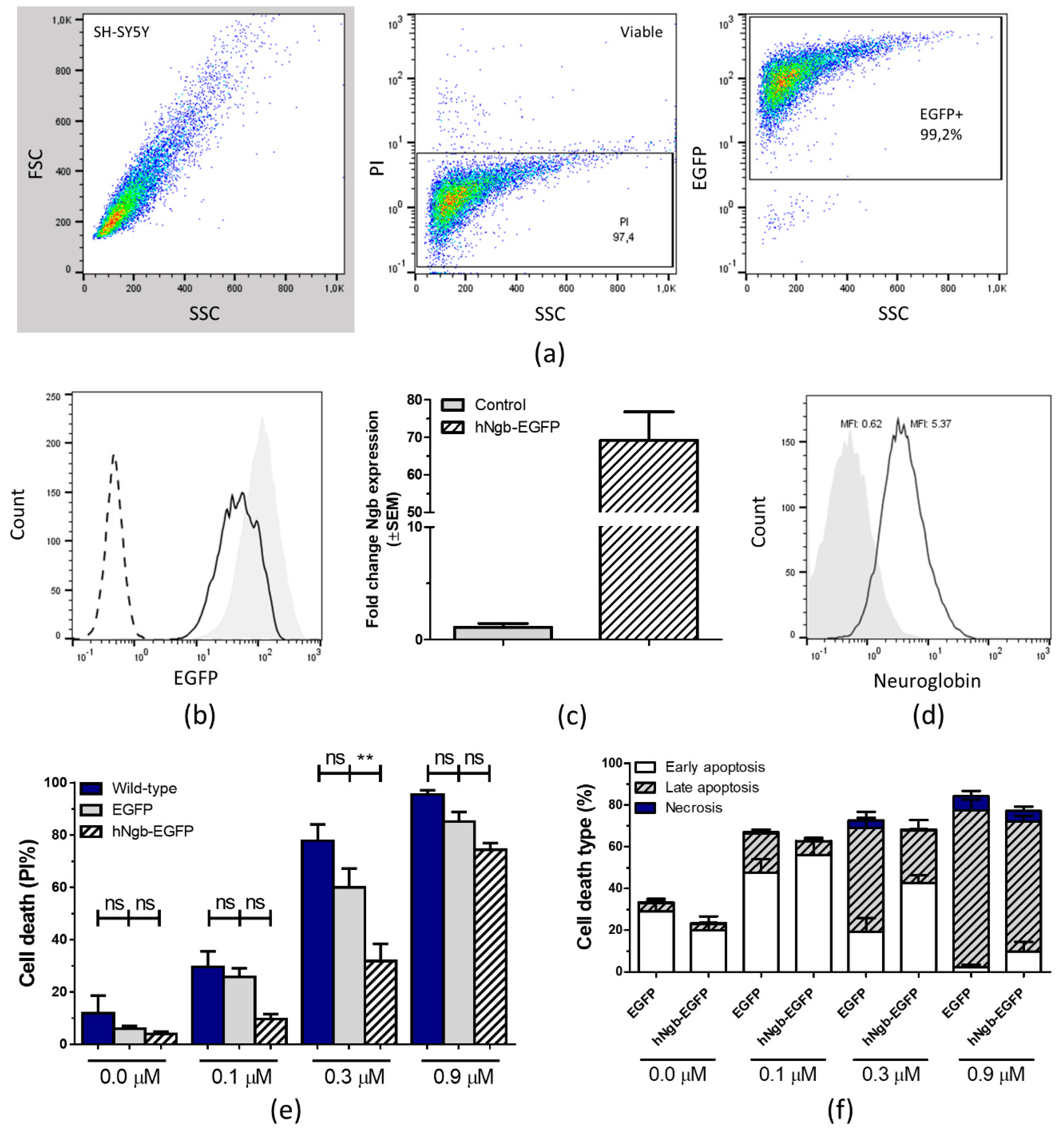
2.2. Flow-Cytometric Phenotyping and Analysis of Cell Viability
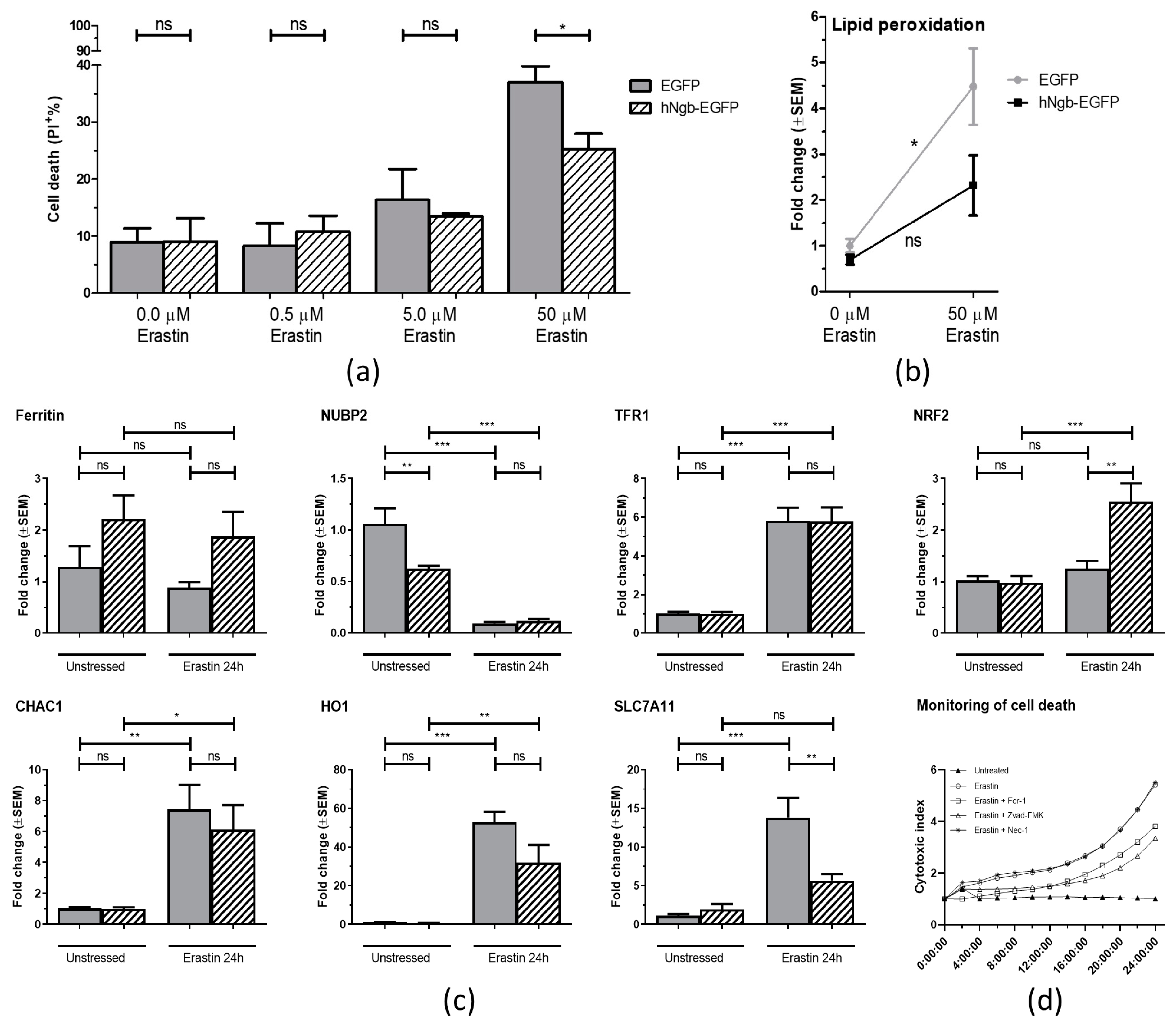
2.3. Lipid Peroxidation
2.4. IncuCyte Kinetic Monitoring of Cell Death
2.5. RNA Extraction and Reverse Transcription Quantitative PCR
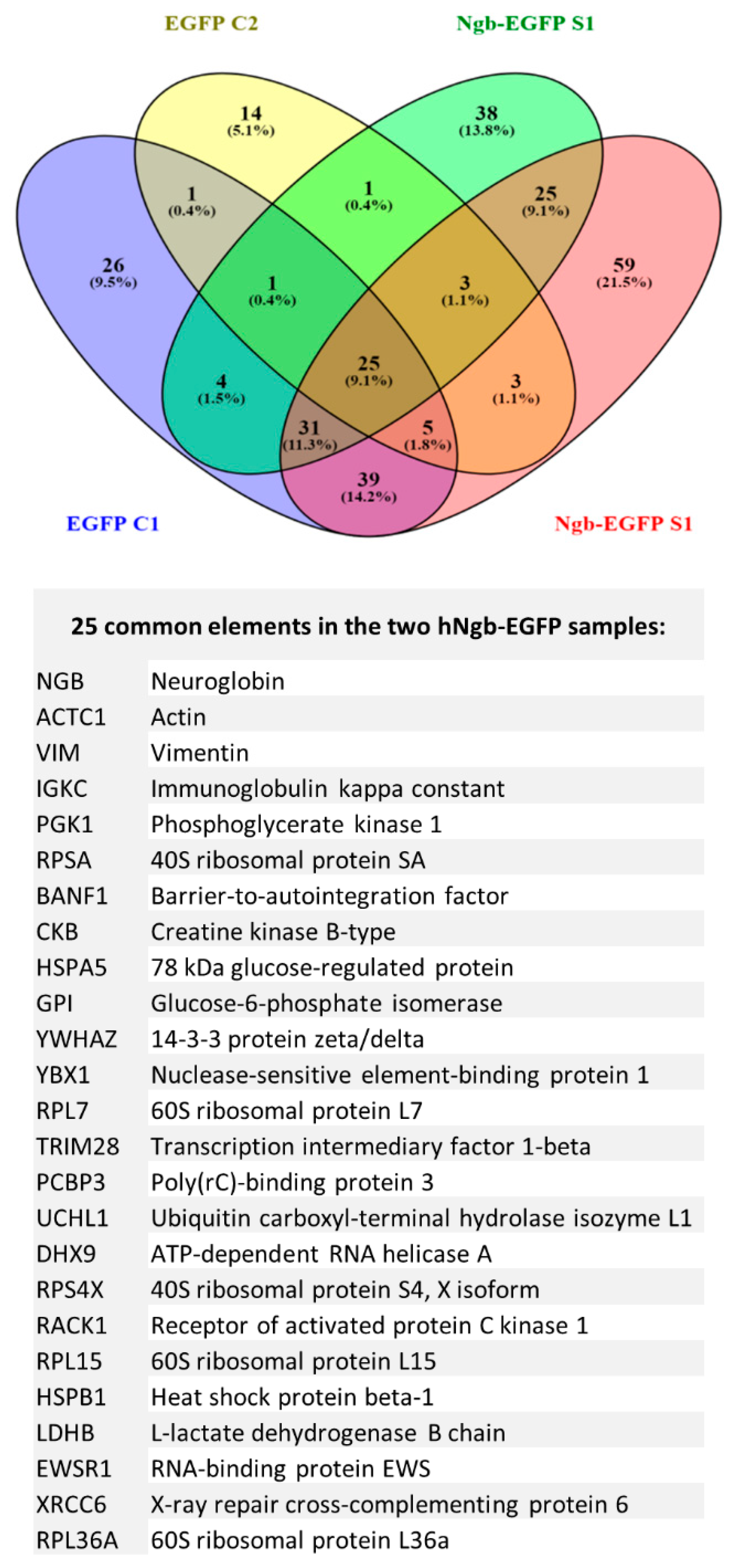
2.6. Protein-Protein Interactions
2.6.1. Cell Culturing
2.6.2. Co-Immunoprecipitation
2.6.3. Protein Digestion
2.6.4. LC-MS/MS Analysis
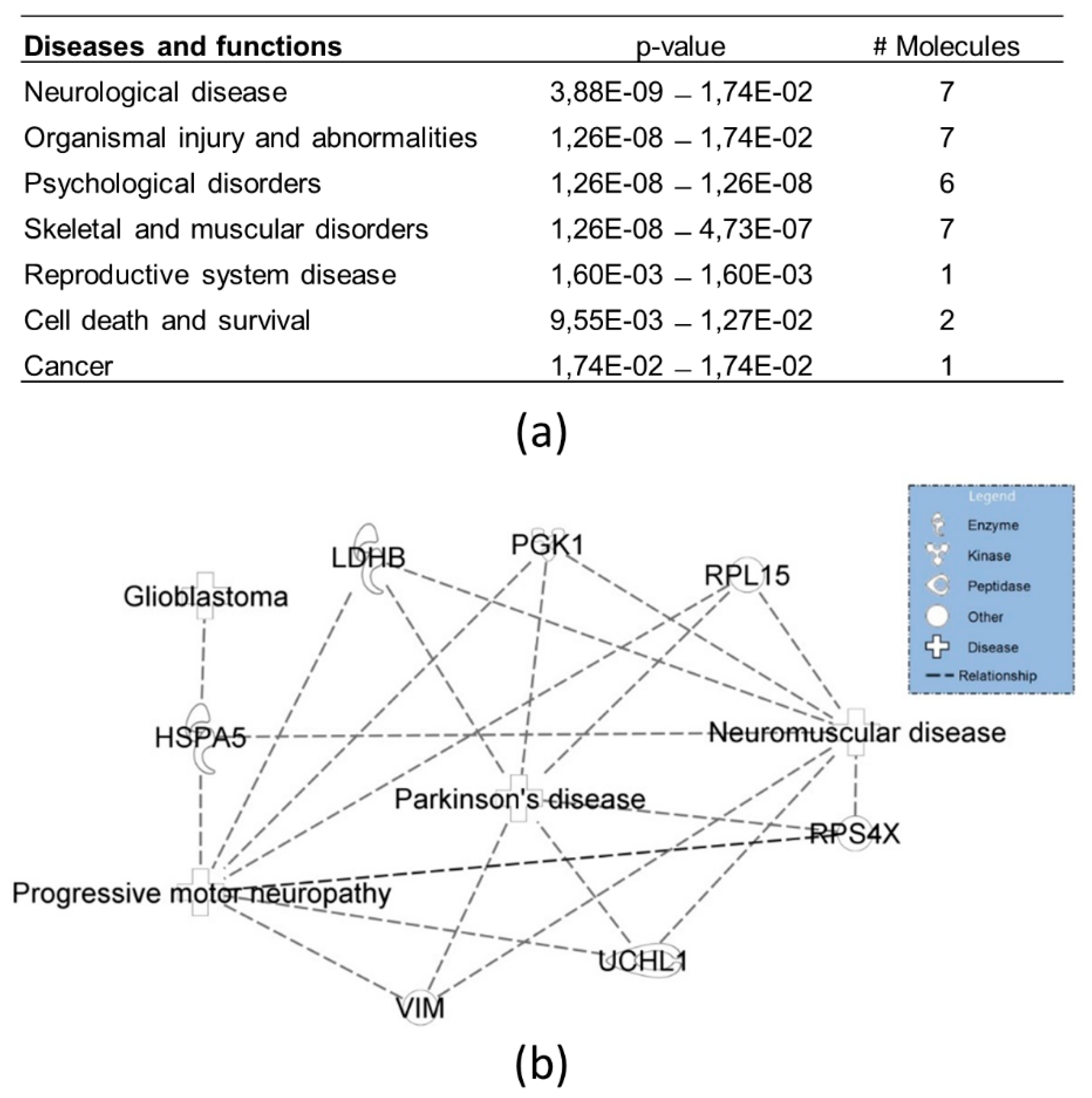
2.6.5. Mass Spectrometric Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Characterization and Validation of a hNgb-EGFP SH-SY5Y Cell Line
3.2. hNgb-EGFP SH-SY5Y Cells Have a Higher Threshold for Ferroptosis Initiation
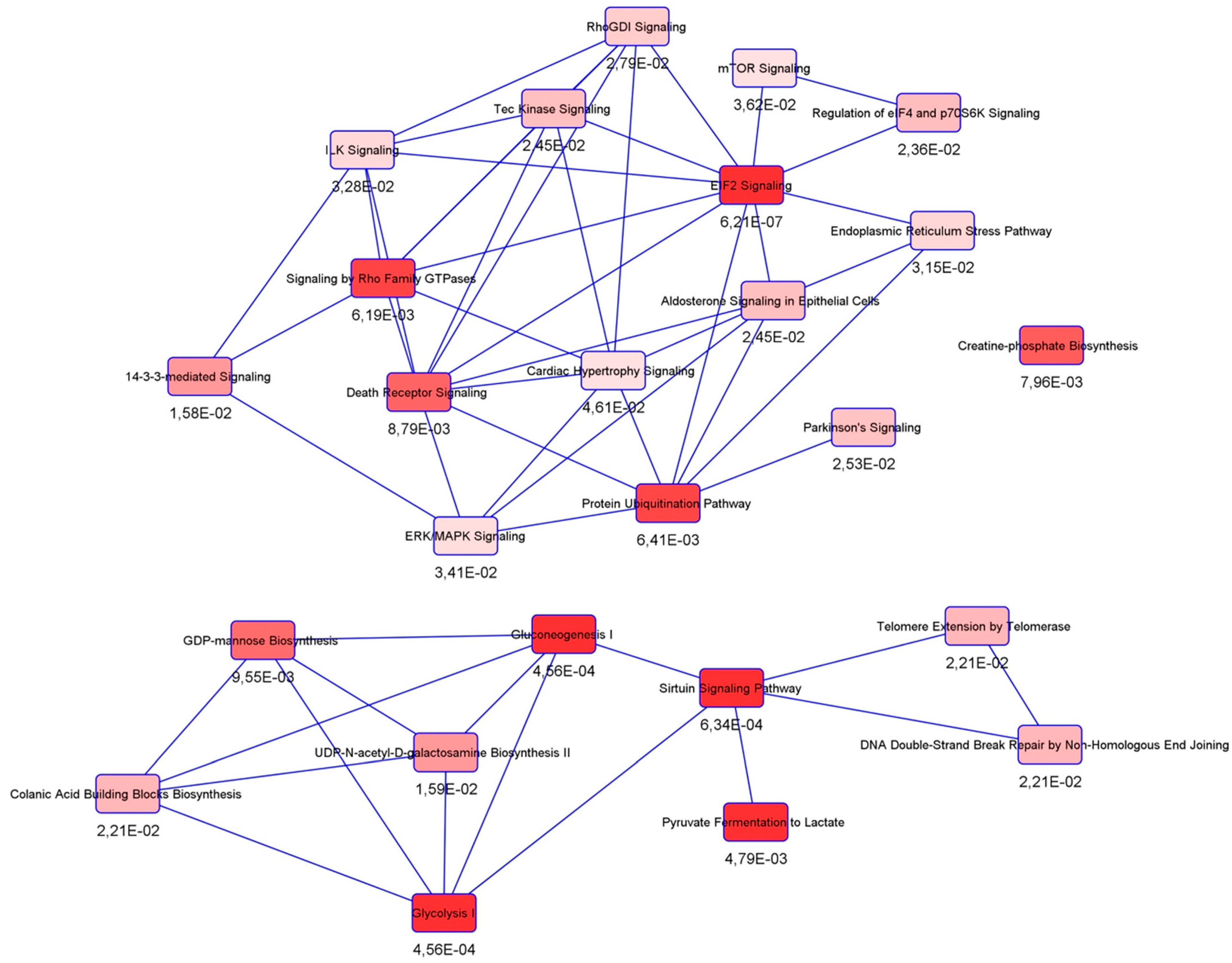
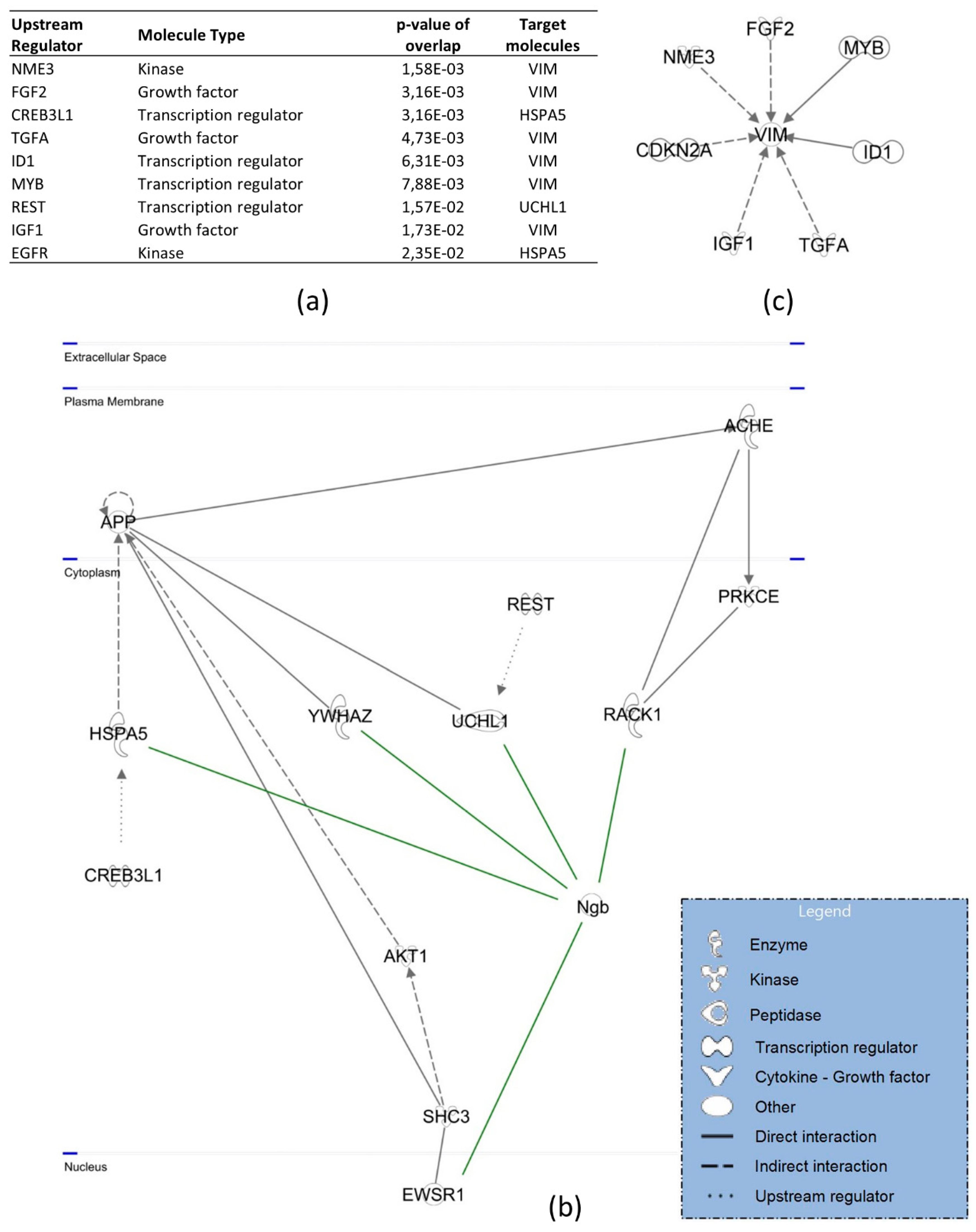
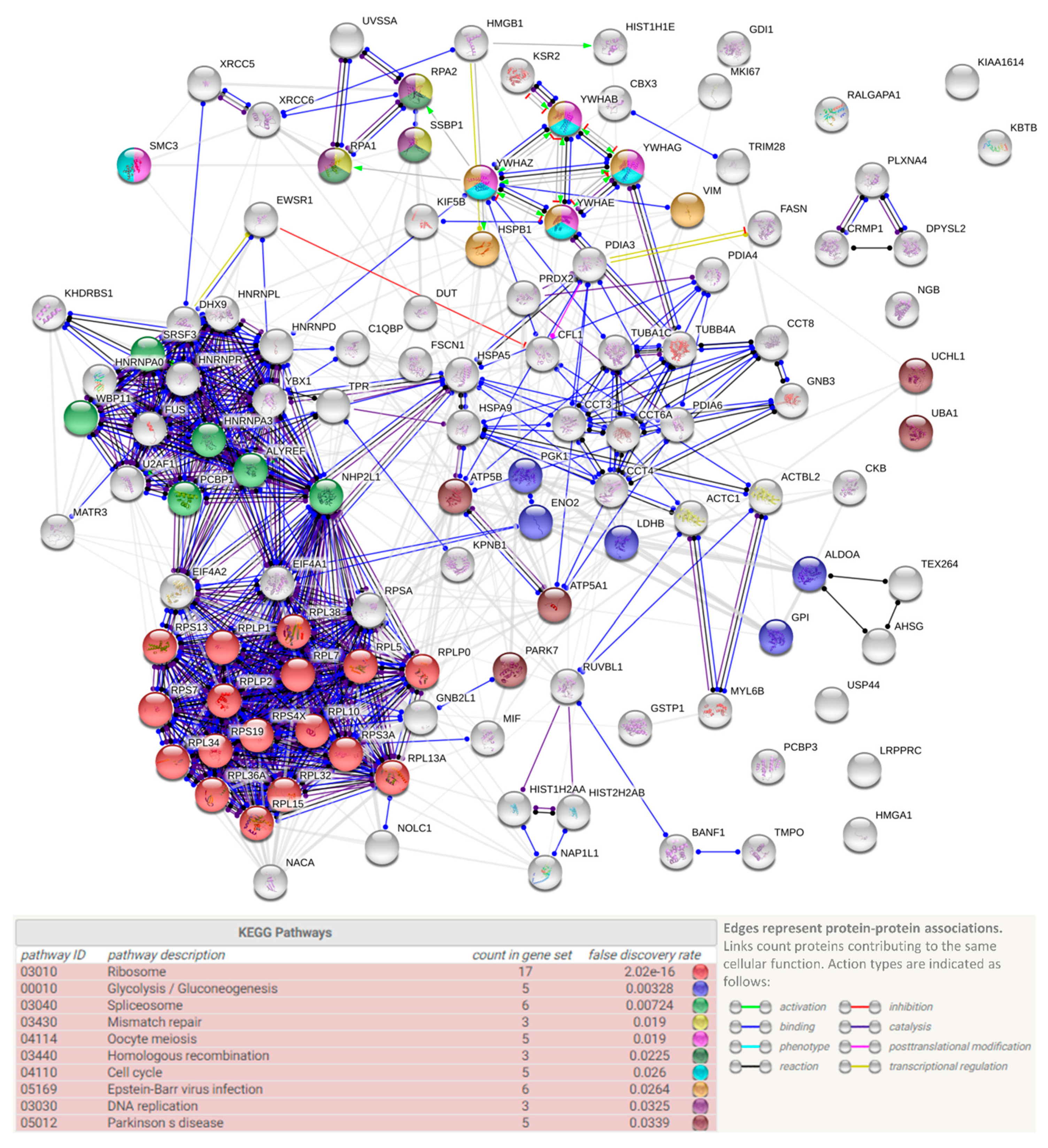
3.3. Protein Interactors of hNgb in a Non-Stress Condition are Enriched in the ‘Cell Death and Survival, Cellular Development, Cellular Growth, and Proliferation’ Network
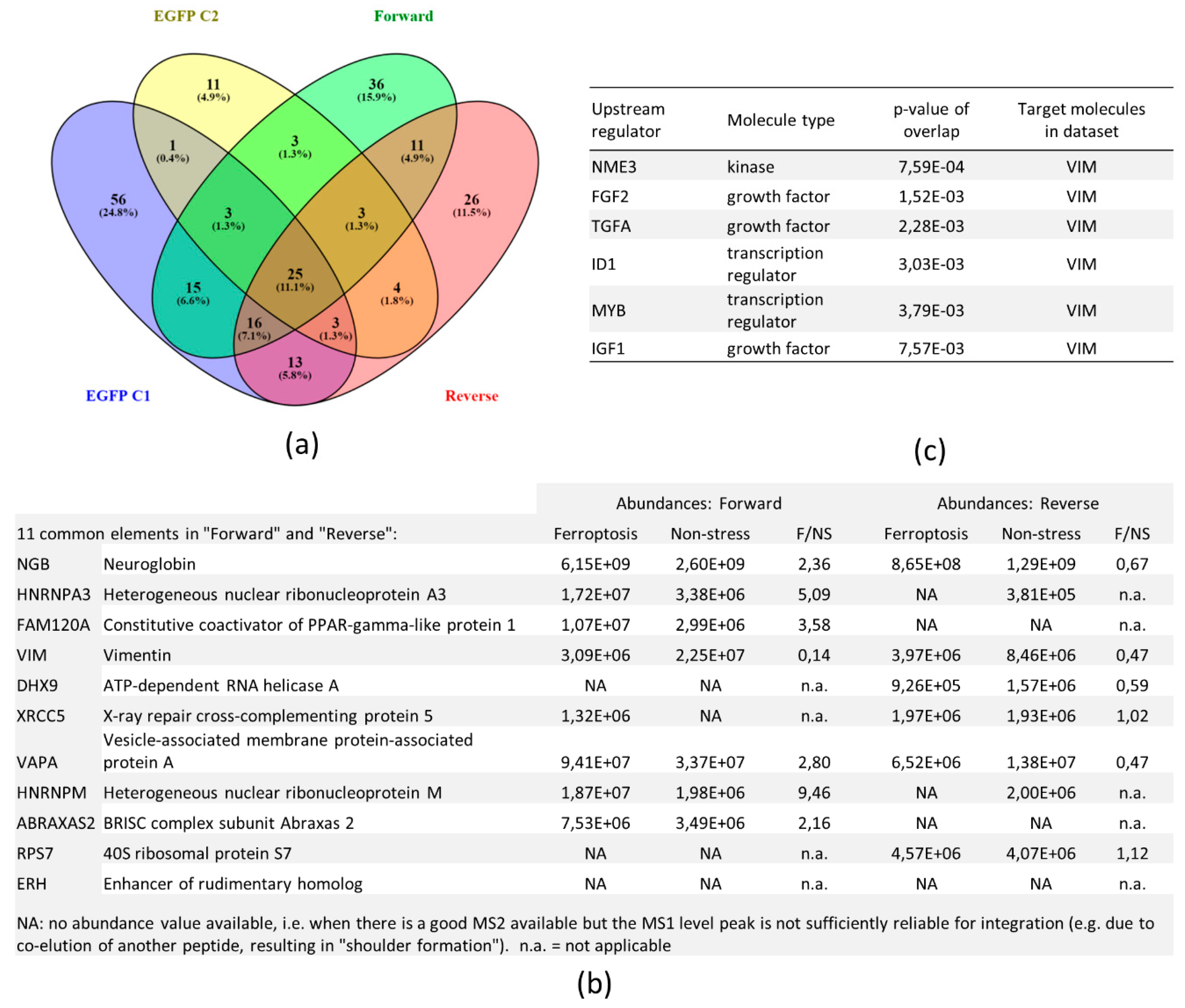
3.4. SILAC Labeling Shows Ferroptosis-Specific hNgb Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Munch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Belaidi, A.A.; Bush, A.I. Iron neurochemistry in Alzheimer’s disease and Parkinson’s disease: Targets for therapeutics. J. Neurochem. 2016, 139, 179–197. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Jomova, K.; Rhodes, C.J.; Kuca, K.; Musilek, K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016, 90, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, D.H.; Choudry, H.A.; Bartlett, D.L.; Lee, Y.J. Ferroptosis-Induced Endoplasmic Reticulum Stress: Cross-talk between Ferroptosis and Apoptosis. Mol. Cancer Res. 2018, 16, 1073–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linkermann, A.; Skouta, R.; Himmerkus, N.; Mulay, S.R.; Dewitz, C.; De Zen, F.; Prokai, A.; Zuchtriegel, G.; Krombach, F.; Welz, P.S.; et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. USA 2014, 111, 16836–16841. [Google Scholar] [CrossRef] [Green Version]
- Hambright, W.S.; Fonseca, R.S.; Chen, L.; Na, R.; Ran, Q. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol. 2017, 12, 8–17. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascon, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Han, X.; Lan, X.; Gao, Y.; Wan, J.; Durham, F.; Cheng, T.; Yang, J.; Wang, Z.; Jiang, C.; et al. Inhibition of neuronal ferroptosis protects hemorrhagic brain. JCI Insight 2017, 2, e90777. [Google Scholar] [CrossRef] [Green Version]
- Tuo, Q.Z.; Lei, P.; Jackman, K.A.; Li, X.L.; Xiong, H.; Liuyang, Z.Y.; Roisman, L.; Zhang, S.T.; Ayton, S.; Wang, Q.; et al. Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol. Psychiatry 2017, 22, 1520–1530. [Google Scholar] [CrossRef]
- Linkermann, A. Nonapoptotic cell death in acute kidney injury and transplantation. Kidney Int. 2016, 89, 46–57. [Google Scholar] [CrossRef] [Green Version]
- Skouta, R.; Dixon, S.J.; Wang, J.; Dunn, D.E.; Orman, M.; Shimada, K.; Rosenberg, P.A.; Lo, D.C.; Weinberg, J.M.; Linkermann, A.; et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 2014, 136, 4551–4556. [Google Scholar] [CrossRef]
- Do Van, B.; Gouel, F.; Jonneaux, A.; Timmerman, K.; Gele, P.; Petrault, M.; Bastide, M.; Laloux, C.; Moreau, C.; Bordet, R.; et al. Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol. Dis. 2016, 94, 169–178. [Google Scholar] [CrossRef]
- Manz, D.H.; Blanchette, N.L.; Paul, B.T.; Torti, F.M.; Torti, S.V. Iron and cancer: Recent insights. Ann. NY Acad. Sci. 2016, 1368, 149–161. [Google Scholar] [CrossRef]
- Dolma, S.; Lessnick, S.L.; Hahn, W.C.; Stockwell, B.R. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 2003, 3, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Yagoda, N.; von Rechenberg, M.; Zaganjor, E.; Bauer, A.J.; Yang, W.S.; Fridman, D.J.; Wolpaw, A.J.; Smukste, I.; Peltier, J.M.; Boniface, J.J.; et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007, 447, 864–868. [Google Scholar] [CrossRef]
- Hassannia, B.; Wiernicki, B.; Ingold, I.; Qu, F.; Van Herck, S.; Tyurina, Y.Y.; Bayir, H.; Abhari, B.A.; Angeli, J.P.F.; Choi, S.M.; et al. Nano-targeted induction of dual ferroptotic mechanisms eradicates high-risk neuroblastoma. J. Clin. Invest. 2018, 128, 3341–3355. [Google Scholar] [CrossRef]
- Van Acker, Z.P.; Luyckx, E.; Dewilde, S. Neuroglobin Expression in the Brain: A Story of Tissue Homeostasis Preservation. Mol. Neurobiol. 2019, 56, 2101–2122. [Google Scholar] [CrossRef]
- Van Acker, Z.P.; Declerck, K.; Luyckx, E.; Vanden Berghe, W.; Dewilde, S. Non-Methylation-Linked Mechanism of REST-Induced Neuroglobin Expression Impacts Mitochondrial Phenotypes in a Mouse Model of Amyotrophic Lateral Sclerosis. Neuroscience 2019, 412, 233–247. [Google Scholar] [CrossRef]
- Van Acker, Z.P.; Luyckx, E.; Van Leuven, W.; Geuens, E.; De Deyn, P.P.; Van Dam, D.; Dewilde, S. Impaired hypoxic tolerance in APP23 mice: A dysregulation of neuroprotective globin levels. FEBS Lett. 2017, 591, 1321–1332. [Google Scholar] [CrossRef]
- Lu, T.; Aron, L.; Zullo, J.; Pan, Y.; Kim, H.; Chen, Y.; Yang, T.H.; Kim, H.M.; Drake, D.; Liu, X.S.; et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature 2014, 507, 448–454. [Google Scholar] [CrossRef]
- Khan, A.A.; Mao, X.O.; Banwait, S.; Jin, K.; Greenberg, D.A. Neuroglobin attenuates beta-amyloid neurotoxicity in vitro and transgenic Alzheimer phenotype in vivo. Proc. Natl. Acad. Sci. USA 2007, 104, 19114–19119. [Google Scholar] [CrossRef]
- Jayaraman, T.; Tejero, J.; Chen, B.B.; Blood, A.B.; Frizzell, S.; Shapiro, C.; Tiso, M.; Hood, B.L.; Wang, X.; Zhao, X.; et al. 14-3-3 binding and phosphorylation of neuroglobin during hypoxia modulate six-to-five heme pocket coordination and rate of nitrite reduction to nitric oxide. J. Biol. Chem. 2011, 286, 42679–42689. [Google Scholar] [CrossRef]
- Fago, A.; Mathews, A.J.; Moens, L.; Dewilde, S.; Brittain, T. The reaction of neuroglobin with potential redox protein partners cytochrome b5 and cytochrome c. FEBS Lett. 2006, 580, 4884–4888. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, N.; Wang, Y.; Li, X.; Wang, X. Identification of neuroglobin-interacting proteins using yeast two-hybrid screening. Neuroscience 2012, 200, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Wakasugi, K.; Nakano, T.; Kitatsuji, C.; Morishima, I. Human neuroglobin interacts with flotillin-1, a lipid raft microdomain-associated protein. Biochem. Biophys. Res. Commun. 2004, 318, 453–460. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef]
- Yamashita, T.; Hafsi, L.; Masuda, E.; Tsujino, H.; Uno, T. Ferric human neuroglobin scavenges superoxide to form oxy adduct. Chem. Pharm. Bull. 2014, 62, 613–615. [Google Scholar] [CrossRef]
- Fordel, E.; Thijs, L.; Martinet, W.; Lenjou, M.; Laufs, T.; Van Bockstaele, D.; Moens, L.; Dewilde, S. Neuroglobin and cytoglobin overexpression protects human SH-SY5Y neuroblastoma cells against oxidative stress-induced cell death. Neurosci. Lett. 2006, 410, 146–151. [Google Scholar] [CrossRef]
- Dixon, S.J.; Patel, D.N.; Welsch, M.; Skouta, R.; Lee, E.D.; Hayano, M.; Thomas, A.G.; Gleason, C.E.; Tatonetti, N.P.; Slusher, B.S.; et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 2014, 3, e02523. [Google Scholar] [CrossRef]
- Adedoyin, O.; Boddu, R.; Traylor, A.; Lever, J.M.; Bolisetty, S.; George, J.F.; Agarwal, A. Heme oxygenase-1 mitigates ferroptosis in renal proximal tubule cells. Am. J. Physiol. Ren. Physiol. 2018, 314, F702–F714. [Google Scholar] [CrossRef] [Green Version]
- Burmester, T.; Weich, B.; Reinhardt, S.; Hankeln, T. A vertebrate globin expressed in the brain. Nature 2000, 407, 520–523. [Google Scholar] [CrossRef]
- Fordel, E.; Thijs, L.; Moens, L.; Dewilde, S. Neuroglobin and cytoglobin expression in mice. Evidence for a correlation with reactive oxygen species scavenging. FEBS J. 2007, 274, 1312–1317. [Google Scholar] [CrossRef]
- Cwerman-Thibault, H.; Lechauve, C.; Augustin, S.; Roussel, D.; Reboussin, E.; Mohammad, A.; Degardin-Chicaud, J.; Simonutti, M.; Liang, H.; Brignole-Baudouin, F.; et al. Neuroglobin Can Prevent or Reverse Glaucomatous Progression in DBA/2J Mice. Mol. Ther. Methods Clin. Dev. 2017, 5, 200–220. [Google Scholar] [CrossRef] [Green Version]
- Baez, E.; Echeverria, V.; Cabezas, R.; Avila-Rodriguez, M.; Garcia-Segura, L.M.; Barreto, G.E. Protection by Neuroglobin Expression in Brain Pathologies. Front. Neurol. 2016, 7, 146. [Google Scholar] [CrossRef]
- Teumer, A.; Qi, Q.; Nethander, M.; Aschard, H.; Bandinelli, S.; Beekman, M.; Berndt, S.I.; Bidlingmaier, M.; Broer, L.; Cappola, A.; et al. Genomewide meta-analysis identifies loci associated with IGF-I and IGFBP-3 levels with impact on age-related traits. Aging Cell 2016, 15, 811–824. [Google Scholar] [CrossRef]
- Van Heemst, D. Insulin, IGF-1 and longevity. Aging Dis. 2010, 1, 147–157. [Google Scholar]
- Zimbone, S.; Monaco, I.; Giani, F.; Pandini, G.; Copani, A.G.; Giuffrida, M.L.; Rizzarelli, E. Amyloid Beta monomers regulate cyclic adenosine monophosphate response element binding protein functions by activating type-1 insulin-like growth factor receptors in neuronal cells. Aging Cell 2018, 17. [Google Scholar] [CrossRef]
- De Nazareth, A.M. Type 2 diabetes mellitus in the pathophysiology of Alzheimer’s disease. Dement. Neuropsychol. 2017, 11, 105–113. [Google Scholar] [CrossRef]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef] [Green Version]
- Hundahl, C.A.; Luuk, H.; Ilmjarv, S.; Falktoft, B.; Raida, Z.; Vikesaa, J.; Friis-Hansen, L.; Hay-Schmidt, A. Neuroglobin-deficiency exacerbates Hif1A and c-FOS response, but does not affect neuronal survival during severe hypoxia in vivo. PLoS ONE 2011, 6, e28160. [Google Scholar] [CrossRef]
- Cabezas, R.; Vega-Vela, N.E.; Gonzalez-Sanmiguel, J.; Gonzalez, J.; Esquinas, P.; Echeverria, V.; Barreto, G.E. PDGF-BB Preserves Mitochondrial Morphology, Attenuates ROS Production, and Upregulates Neuroglobin in an Astrocytic Model Under Rotenone Insult. Mol. Neurobiol. 2017. [Google Scholar] [CrossRef]
- Baez-Jurado, E.; Vega, G.G.; Aliev, G.; Tarasov, V.V.; Esquinas, P.; Echeverria, V.; Barreto, G.E. Blockade of Neuroglobin Reduces Protection of Conditioned Medium from Human Mesenchymal Stem Cells in Human Astrocyte Model (T98G) Under a Scratch Assay. Mol. Neurobiol. 2017. [Google Scholar] [CrossRef]
- Luyckx, E.; Everaert, B.R.; Van der Veken, B.; Van Leuven, W.; Timmermans, J.P.; Vrints, C.J.; De Meyer, G.R.Y.; Martinet, W.; Dewilde, S. Cytoprotective effects of transgenic neuroglobin overexpression in an acute and chronic mouse model of ischemic heart disease. Heart Vessel. 2017. [Google Scholar] [CrossRef]
- Zhu, S.; Ding, S.; Wang, P.; Wei, Z.; Pan, W.; Palm, N.W.; Yang, Y.; Yu, H.; Li, H.B.; Wang, G.; et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 2017, 546, 667–670. [Google Scholar] [CrossRef]
- Zeqiraj, E.; Tian, L.; Piggott, C.A.; Pillon, M.C.; Duffy, N.M.; Ceccarelli, D.F.; Keszei, A.F.; Lorenzen, K.; Kurinov, I.; Orlicky, S.; et al. Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function. Mol. Cell 2015, 59, 970–983. [Google Scholar] [CrossRef]
- Levin, E.C.; Acharya, N.K.; Sedeyn, J.C.; Venkataraman, V.; D’Andrea, M.R.; Wang, H.Y.; Nagele, R.G. Neuronal expression of vimentin in the Alzheimer’s disease brain may be part of a generalized dendritic damage-response mechanism. Brain Res. 2009, 1298, 194–207. [Google Scholar] [CrossRef]
- Ververis, A.; Christodoulou, A.; Christoforou, M.; Kamilari, C.; Lederer, C.W.; Santama, N. A novel family of katanin-like 2 protein isoforms (KATNAL2), interacting with nucleotide-binding proteins Nubp1 and Nubp2, are key regulators of different MT-based processes in mammalian cells. Cell. Mol. Life Sci. 2016, 73, 163–184. [Google Scholar] [CrossRef]
- Haines, B.; Mao, X.; Xie, L.; Spusta, S.; Zeng, X.; Jin, K.; Greenberg, D.A. Neuroglobin expression in neurogenesis. Neurosci. Lett. 2013, 549, 3–6. [Google Scholar] [CrossRef]
- Fabrizius, A.; Andre, D.; Laufs, T.; Bicker, A.; Reuss, S.; Burmester, T.; Hankeln, T. Critical re-evaluation of neuroglobin expression reveals conserved patterns among mammals. Neuroscience 2016. [Google Scholar] [CrossRef]
- Li, L.; Liu, Q.R.; Xiong, X.X.; Liu, J.M.; Lai, X.J.; Cheng, C.; Pan, F.; Chen, Y.; Yu, S.B.; Yu, A.C.; et al. Neuroglobin promotes neurite outgrowth via differential binding to PTEN and Akt. Mol. Neurobiol. 2014, 49, 149–162. [Google Scholar] [CrossRef]
- Taylor, J.M.; Kelley, B.; Gregory, E.J.; Berman, N.E. Neuroglobin overexpression improves sensorimotor outcomes in a mouse model of traumatic brain injury. Neurosci. Lett. 2014, 577, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Bentmann, A.; Schmidt, M.; Reuss, S.; Wolfrum, U.; Hankeln, T.; Burmester, T. Divergent distribution in vascular and avascular mammalian retinae links neuroglobin to cellular respiration. J. Biol. Chem. 2005, 280, 20660–20665. [Google Scholar] [CrossRef]
- Lechauve, C.; Augustin, S.; Cwerman-Thibault, H.; Bouaita, A.; Forster, V.; Celier, C.; Rustin, P.; Marden, M.C.; Sahel, J.A.; Corral-Debrinski, M. Neuroglobin involvement in respiratory chain function and retinal ganglion cell integrity. Biochim. Biophys. Acta 2012, 1823, 2261–2273. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, T.; Ferri, A.; Sorice, M.; Azmoon, P.; Grasso, M.; Mattei, V.; Capozzi, A.; Manganelli, V.; Misasi, R. Neuroglobin overexpression plays a pivotal role in neuroprotection through mitochondrial raft-like microdomains in neuroblastoma SK-N-BE2 cells. Mol. Cell Neurosci. 2018, 88, 167–176. [Google Scholar] [CrossRef]
- Watanabe, S.; Takahashi, N.; Uchida, H.; Wakasugi, K. Human neuroglobin functions as an oxidative stress-responsive sensor for neuroprotection. J. Biol. Chem. 2012, 287, 30128–30138. [Google Scholar] [CrossRef]
- Abbruzzetti, S.; Faggiano, S.; Bruno, S.; Spyrakis, F.; Mozzarelli, A.; Dewilde, S.; Moens, L.; Viappiani, C. Ligand migration through the internal hydrophobic cavities in human neuroglobin. Proc. Natl. Acad. Sci. USA 2009, 106, 18984–18989. [Google Scholar] [CrossRef] [Green Version]
- De Maio, A.; Cauvi, D.M.; Capone, R.; Bello, I.; Egberts, W.V.; Arispe, N.; Boelens, W. The small heat shock proteins, HSPB1 and HSPB5, interact differently with lipid membranes. Cell Stress Chaperones 2019. [Google Scholar] [CrossRef]
- Sun, X.; Ou, Z.; Xie, M.; Kang, R.; Fan, Y.; Niu, X.; Wang, H.; Cao, L.; Tang, D. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 2015, 34, 5617–5625. [Google Scholar] [CrossRef] [Green Version]
- Leidgens, S.; Bullough, K.Z.; Shi, H.; Li, F.; Shakoury-Elizeh, M.; Yabe, T.; Subramanian, P.; Hsu, E.; Natarajan, N.; Nandal, A.; et al. Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J. Biol. Chem. 2013, 288, 17791–17802. [Google Scholar] [CrossRef]
- Kurz, T.; Gustafsson, B.; Brunk, U.T. Intralysosomal iron chelation protects against oxidative stress-induced cellular damage. FEBS J. 2006, 273, 3106–3117. [Google Scholar] [CrossRef]
- Thompson, K.; Menzies, S.; Muckenthaler, M.; Torti, F.M.; Wood, T.; Torti, S.V.; Hentze, M.W.; Beard, J.; Connor, J. Mouse brains deficient in H-ferritin have normal iron concentration but a protein profile of iron deficiency and increased evidence of oxidative stress. J. Neurosci. Res. 2003, 71, 46–63. [Google Scholar] [CrossRef]
- Mitsuishi, Y.; Taguchi, K.; Kawatani, Y.; Shibata, T.; Nukiwa, T.; Aburatani, H.; Yamamoto, M.; Motohashi, H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012, 22, 66–79. [Google Scholar] [CrossRef]
- Kerns, M.L.; Hakim, J.M.; Lu, R.G.; Guo, Y.; Berroth, A.; Kaspar, R.L.; Coulombe, P.A. Oxidative stress and dysfunctional NRF2 underlie pachyonychia congenita phenotypes. J. Clin. Investig. 2016, 126, 2356–2366. [Google Scholar] [CrossRef] [Green Version]
- Papp, D.; Lenti, K.; Modos, D.; Fazekas, D.; Dul, Z.; Turei, D.; Foldvari-Nagy, L.; Nussinov, R.; Csermely, P.; Korcsmaros, T. The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops. FEBS Lett. 2012, 586, 1795–1802. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.M.; Xiong, Y.S.; Kong, F.L.; Qu, M.; Wang, Q.; Chen, X.Q.; Wang, J.Z.; Zhu, L.Q. Neuroglobin attenuates Alzheimer-like tau hyperphosphorylation by activating Akt signaling. J. Neurochem. 2012, 120, 157–164. [Google Scholar] [CrossRef]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Endoplasmic reticulum and the unfolded protein response: Dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 2013, 301, 215–290. [Google Scholar] [CrossRef]
- Chen, S.; Lu, F.F.; Seeman, P.; Liu, F. Quantitative proteomic analysis of human substantia nigra in Alzheimer’s disease, Huntington’s disease and Multiple sclerosis. Neurochem. Res. 2012, 37, 2805–2813. [Google Scholar] [CrossRef]
- Filareti, M.; Luotti, S.; Pasetto, L.; Pignataro, M.; Paolella, K.; Messina, P.; Pupillo, E.; Filosto, M.; Lunetta, C.; Mandrioli, J.; et al. Decreased Levels of Foldase and Chaperone Proteins Are Associated with an Early-Onset Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2017, 10, 99. [Google Scholar] [CrossRef]
- Costes, S.; Gurlo, T.; Rivera, J.F.; Butler, P.C. UCHL1 deficiency exacerbates human islet amyloid polypeptide toxicity in beta-cells: Evidence of interplay between the ubiquitin/proteasome system and autophagy. Autophagy 2014, 10, 1004–1014. [Google Scholar] [CrossRef]
- Groh, N.; Buhler, A.; Huang, C.; Li, K.W.; van Nierop, P.; Smit, A.B.; Fandrich, M.; Baumann, F.; David, D.C. Age-Dependent Protein Aggregation Initiates Amyloid-beta Aggregation. Front. Aging Neurosci. 2017, 9, 138. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Acker, Z.P.; Van Raemdonck, G.A.; Logie, E.; Van Acker, S.I.; Baggerman, G.; Vanden Berghe, W.; Ponsaerts, P.; Dewilde, S. Connecting the Dots in the Neuroglobin-Protein Interaction Network of an Unstressed and Ferroptotic Cell Death Neuroblastoma Model. Cells 2019, 8, 873. https://doi.org/10.3390/cells8080873
Van Acker ZP, Van Raemdonck GA, Logie E, Van Acker SI, Baggerman G, Vanden Berghe W, Ponsaerts P, Dewilde S. Connecting the Dots in the Neuroglobin-Protein Interaction Network of an Unstressed and Ferroptotic Cell Death Neuroblastoma Model. Cells. 2019; 8(8):873. https://doi.org/10.3390/cells8080873
Chicago/Turabian StyleVan Acker, Zoë P., Geert A. Van Raemdonck, Emilie Logie, Sara I. Van Acker, Geert Baggerman, Wim Vanden Berghe, Peter Ponsaerts, and Sylvia Dewilde. 2019. "Connecting the Dots in the Neuroglobin-Protein Interaction Network of an Unstressed and Ferroptotic Cell Death Neuroblastoma Model" Cells 8, no. 8: 873. https://doi.org/10.3390/cells8080873




