An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Renal Histology and Immunohistochemistry
2.3. Exosome and Pellet Purification and Characterisation
2.4. RNA Isolation from Urinary Cell Pellet and Exosomes
2.5. Individual Assay qPCR-RT
2.6. Pathway Analysis
2.7. Immunofluorescence in Renal Biopsy
2.8. Human Kidney Cells Culture
2.9. Overexpression of miR-21/miR-150 and Inhibition of mir-29c in Human Kidney Cells
2.10. Luciferase Activity Analysis
2.11. Statistical Analysis
3. Results
3.1. Patients
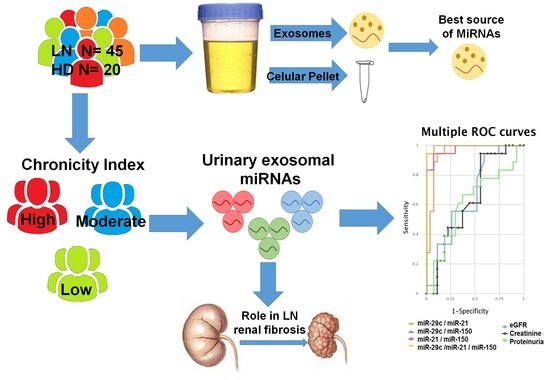
3.2. Isolation and Characterization of Urinary Exosomes and Cellular Pellet
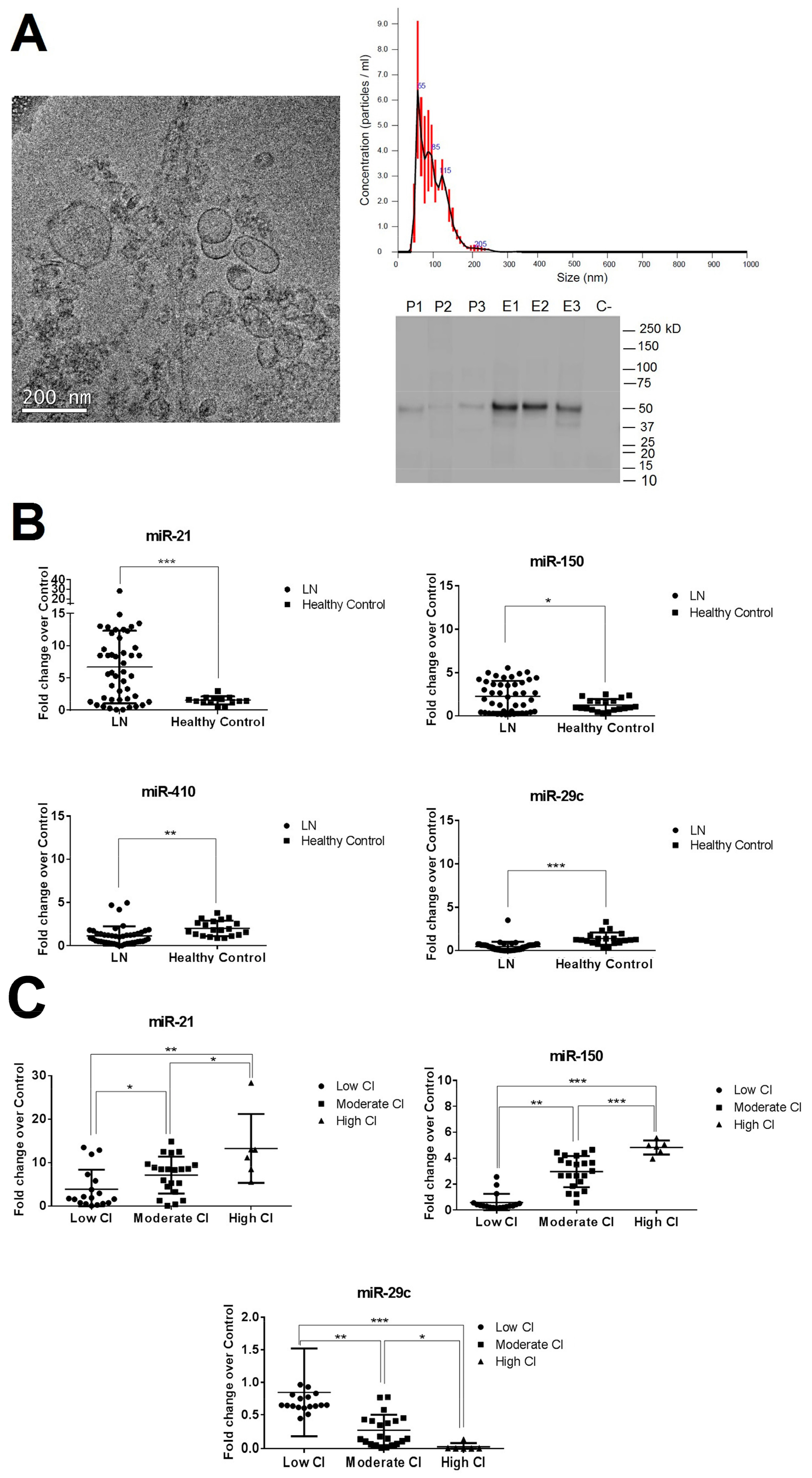
3.3. miR-29c, miR-200a, miR-21, miR-410, and miR-150 Expression Levels Measured by Quantitative Reverse Transcription-Polymerase Chain Reaction (qPCR-RT) in Urinary Exosomal Preparations and Cellular Pellet
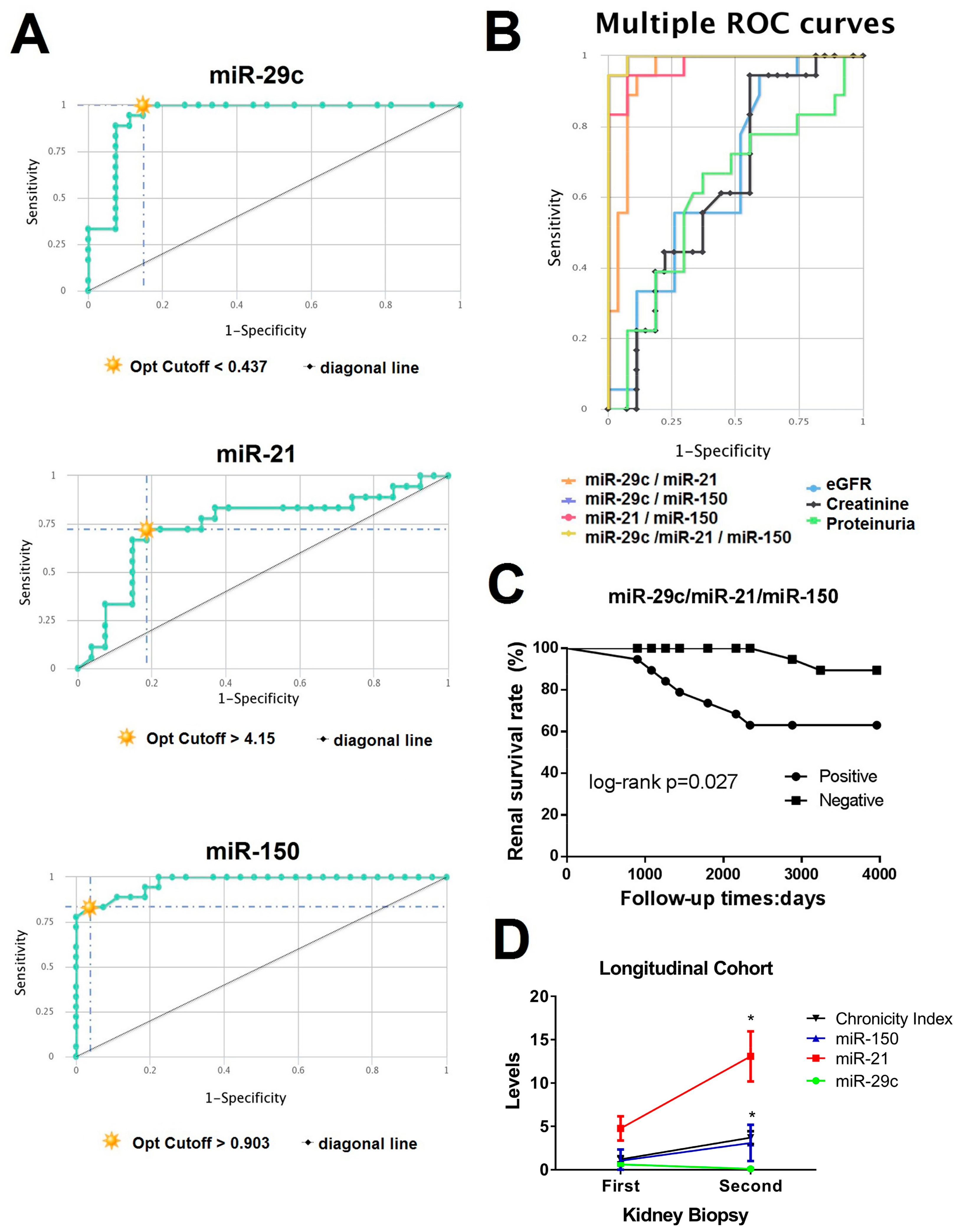
3.4. CombiROC Performance Analyses for Optimal Marker Combinations
3.5. Pathway Enrichment Analyses
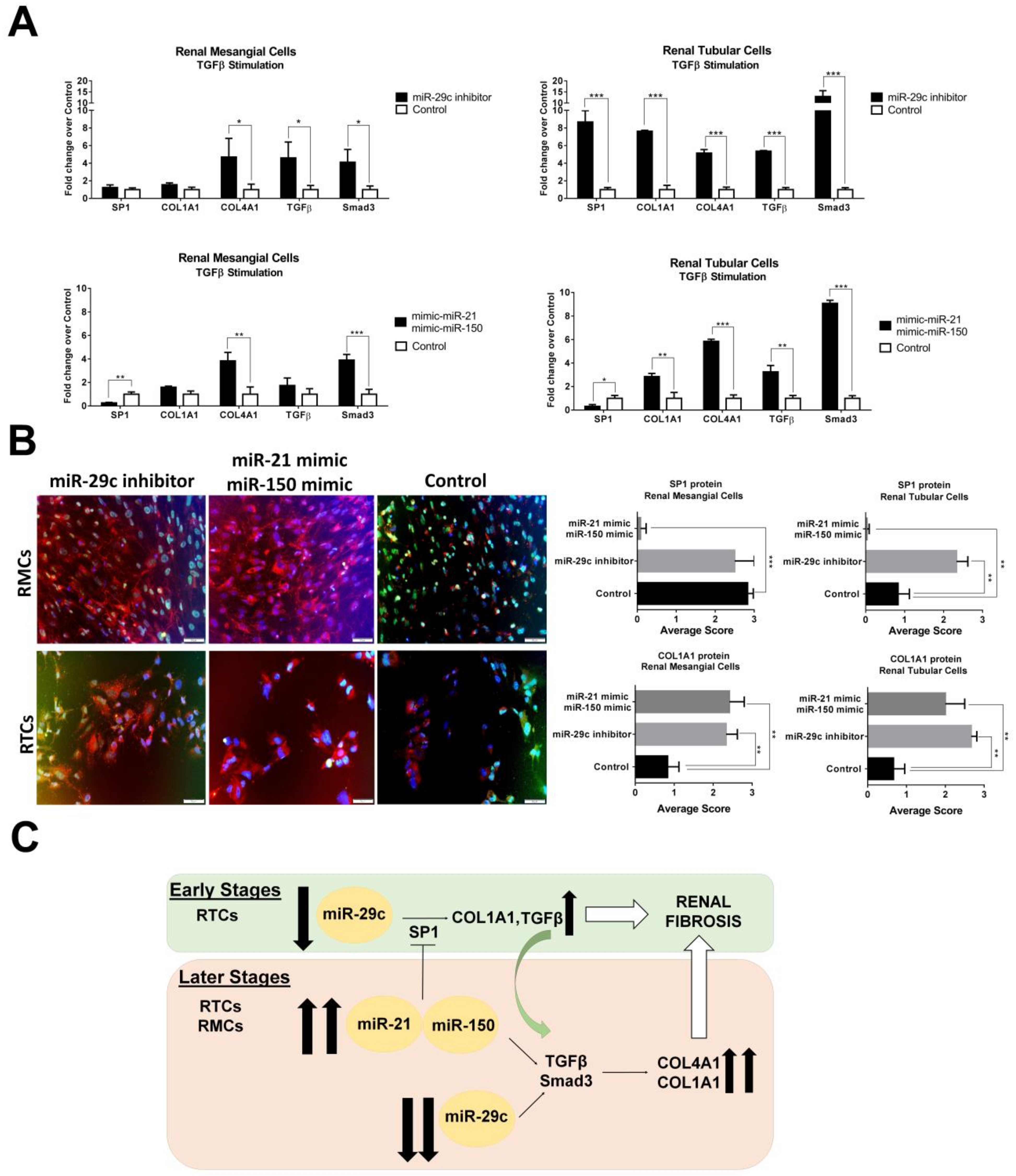
3.6. Confirmation of SP1 as Common Urinary miRNAs Target and Associated Profibrotic Molecules in Kidney Biopsies
3.7. Over-Expression of miR-21-5p/miR-150 and Inhibition of miR-29c Increase Profibrotic Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boor, P.; Ostendorf, T.; Floege, J. Renal fibrosis: Novel insights into mechanisms and therapeutic targets. Nat. Rev. Nephrol. 2010, 6, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Sheppard, D.; Duffield, J.S.; Violette, S. Therapy for fibrotic diseases: Nearing the starting line. Sci. Transl. Med. 2013, 5, 167sr1. [Google Scholar] [CrossRef]
- Pottier, N.; Cauffiez, C.; Perrais, M.; Barbry, P.; Mari, B. FibromiRs: Translating molecular discoveries into new anti-fibrotic drugs. Trends Pharmacol. Sci. 2014, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C.; Lo, M.S.; Costa Reis, P.; Sullivan, K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2016, 12, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Almaani, S.; Meara, A.; Rovin, B.H. Update on Lupus Nephritis. Clin. J. Am. Soc. Nephrol. 2017, 12, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Moroni, G.; Depetri, F.; Ponticelli, C. Lupus nephritis: When and how often to biopsy and what does it mean? J. Autoimmun. 2016, 74, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A.F. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Kwan, B.C.; Lai, F.M.; Chow, K.M.; Kam-Tao, L.P.; Szeto, C.C. Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis. Markers 2010, 28, 79–86. [Google Scholar] [CrossRef]
- Miah, S.; Dudziec, E.; Drayton, R.M.; Zlotta, A.R.; Morgan, S.L.; Rosario, D.J.; Hamdy, F.C.; Catto, J.W. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br. J. Cancer 2012, 107, 123–128. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colan, R.S.; Pisano, S.; Oliveira, M.I.; Ferrari, M.; Mendes Pinto, I. Exosomes as reconfigurable therapeutic systems. Trends Mol. Med. 2017, 23, 636–650. [Google Scholar] [CrossRef] [PubMed]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef]
- Santiago-Dieppa, D.R.; Steinberg, J.; Gonda, D.; Cheung, V.J.; Carter, B.S.; Chen, C.C. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev. Mol. Diagn. 2014, 14, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Zhao, Z.; Yang, T.; Wang, Y.; Li, B.; Lv, J.; Ding, W. Liquid biopsy biomarkers of renal interstitial fibrosis based on urinary exosome. Exp. Mol. Pathol. 2018, 105, 223–228. [Google Scholar] [CrossRef]
- Zarjou, A.; Yang, S.; Abraham, E.; Agarwal, A.; Liu, G. Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am. J. Physiol. Ren. Physiol. 2011, 301, F793–F801. [Google Scholar] [CrossRef]
- Rudnicki, M.; Perco, P.; Leierer, J.; Heinzel, A.; Mühlberger, I.; Schweibert, N.; Sunzenauer, J.; Regele, H.; Kronbichler, A.; Mestdagh, P. Renal microRNA-and RNA-profiles in progressive chronic kidney disease. Eur. J. Clin. Investig. 2016, 46, 213–226. [Google Scholar] [CrossRef]
- Muralidharan, J.; Ramezani, A.; Hubal, M.; Knoblach, S.; Shrivastav, S.; Karandish, S.; Scott, R.; Maxwell, N.; Ozturk, S.; Beddhu, S. Extracellular microRNA signature in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2017, 312, F982–F991. [Google Scholar] [CrossRef] [Green Version]
- Gholaminejad, A.; Abdul Tehrani, H.; Gholami Fesharaki, M. Identification of candidate microRNA biomarkers in renal fibrosis: A meta-analysis of profiling studies. Biomarkers 2018, 23, 713–724. [Google Scholar] [CrossRef]
- Lv, L.L.; Cao, Y.H.; Ni, H.F.; Xu, M.; Liu, D.; Liu, H.; Chen, P.S.; Liu, B.C. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am. J. Physiol. Ren. Physiol. 2013, 305, F1220–F1227. [Google Scholar] [CrossRef] [Green Version]
- Lv, C.Y.; Ding, W.J.; Wang, Y.L.; Zhao, Z.Y.; Li, J.H.; Chen, Y.; Lv, J. A PEG-based method for the isolation of urinary exosomes and its application in renal fibrosis diagnostics using cargo miR-29c and miR-21 analysis. Int. Urol. Nephrol. 2018, 50, 973–982. [Google Scholar] [CrossRef]
- Yu, Y.; Bai, F.; Qin, N.; Liu, W.; Sun, Q.; Zhou, Y.; Yang, J. Non-proximal renal tubule-derived urinary exosomal miR-200b as a biomarker of renal fibrosis. Nephron 2018, 139, 269–282. [Google Scholar] [CrossRef]
- Meng, X.M.; Tang, P.M.; Li, J.; Lan, H.Y. TGF-β/Smad signaling in renal fibrosis. Front. Physiol. 2015, 6, 82. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, N.; Zhang, J.; Zhao, H.; Wang, X. miR-410 suppresses the expression of interleukin-6 as well as renal fibrosis in the pathogenesis of lupus nephritis. Clin. Exp. Pharmacol. Physiol. 2016, 43, 616–625. [Google Scholar] [CrossRef]
- Solé, C.; Cortés-Hernández, J.; Felip, M.L.; Vidal, M.; Ordi-Ros, J. MiR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol. Dial. Transplant. 2015, 30, 1488–1496. [Google Scholar] [CrossRef]
- Zhou, H.; Hasni, S.A.; Perez, P.; Tandom, M.; Jang, S.I.; Zheng, C.; Kopp, J.B.; Austin, H., 3rd; Balow, J.E.; Alevizos, I.; et al. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J. Am. Soc. Nephrol. 2013, 24, 1073–1087. [Google Scholar] [CrossRef]
- Kim, B.K.; Lee, J.W.; Park, P.J.; Shin, Y.S.; Lee, W.Y.; Lee, K.A.; Ye, S.; Hyun, H.; Kang, K.N.; Yeo, D.; et al. The multiplex bead array approach to identifying serum biomarkers associated with breast cancer. Breast Cancer Res. 2009, 11, R22. [Google Scholar] [CrossRef]
- Freydanck, M.K.; Laubender, R.P.; Rack, B.; Shuhmacher, L.; Jeschke, U.; Scholz, C. Two-marker combinations for preoperative discrimination of benign and malignant ovarian masses. Anticancer Res. 2012, 32, 2003–2008. [Google Scholar] [PubMed]
- Panebianco, F.; Mazzanti, C.; Tomei, S.; Aretini, P.; Franceschi, S.; Lessi, F.; Di Coscio, G.; Bevilacgua, G.; Marchetti, I. The combination of four molecular markers improves thyroid cancer cytologic diagnosis and patient management. BMC Cancer 2015, 15, 918. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American college of rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef]
- Gladman, D.D.; Ibanez, D.; Urowitz, M.B. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 2002, 29, 288–291. [Google Scholar] [PubMed]
- Coresh, J.; Turin, T.C.; Matsushita, K.; Sang, Y.; Ballew, S.H.; Appel, L.J.; Arima, H.; Chadban, S.J.; Cirillo, M.; Djurdjev, O.; et al. Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA 2014, 311, 2518–2531. [Google Scholar] [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Weening, J.J.; D’Agati, V.D.; Schwartz, M.M.; Seshan, S.V.; Alpers, C.E.; Appel, G.B.; Balow, J.E.; Brujin, J.A.; Cook, T.; Ferrario, F.; et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J. Am. Soc. Nephrol. 2004, 15, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Austin, H.A., 3rd; Muenz, L.R.; Joyce, K.M.; Antonovych, T.A.; Kullick, M.E.; Klippel, J.H.; Decker, J.L.; Balow, J.E. Prognostic factors in lupus nephritis. Contribution of renal histologic data. Am. J. Med. 1983, 75, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef]
- Fan, Y.; Xia, J. MiRNet-Functional Analysis and Visual Exploration of miRNA-Target Interactions in a Network Context. Methods Mol. Biol. 2018, 1819, 215–233. [Google Scholar] [CrossRef]
- Mason, D.Y.; Micklem, K.; Jones, M. Double immunofluorescence labelling of routinely processed paraffin sections. J. Pathol. 2000, 141, 452–461. [Google Scholar] [CrossRef]
- Mazzara, S.; Rossi, R.L.; Grifantini, R.; Donizetti, S.; Abrignani, S.; Bombaci, M. CombiROC: An interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 2017, 7, 45477. [Google Scholar] [CrossRef]
- Spencer, J.D.; Schwaderer, A.L.; Dirosario, J.D.; McHugh, K.M.; McGillivary, G.; Justice, S.S.; Carpenter, A.R.; Baker, P.B.; Harder, J.; Hains, D.S. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011, 80, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef]
- Zingaretti, C.; Arigò, M.; Cardaci, A.; Moro, M.; Crosti, M.; Sinisi, A.; Sugliano, E.; Cheroni, C.; Marabita, F.; Nogarotto, R.; et al. Identification of New Autoantigens by Protein Array Indicates a Role for IL4 Neutralization in Autoimmune Hepatitis. Mol. Cell. Proteom. 2012, 11, 1885–1897. [Google Scholar] [CrossRef] [Green Version]
- Cano-Rodriguez, D.; Campagnoli, S.; Grandi, A.; Parri, M.; Camili, E.; Song, C.; Jin, B.; Lacombe, A.; Pierleoni, A.; Bombaci, M.; et al. TCTN2: A novel tumor marker with oncogenic properties. Oncotarget 2017, 8, 95256–95269. [Google Scholar] [CrossRef]
- Rathnayake, D.; Chang, T.; Udagama, P. Selected serum cytokines and nitric oxide as potential multi-marker biosignature panels for Parkinson disease of varying durations: A case-control study. BMC Neurol. 2019, 19, 56. [Google Scholar] [CrossRef]
- Yung, S.; Yap, D.Y.H.; Chan, T.M. Recent advances in the understanding of renal inflammation and fibrosis in lupus nephritis. F1000Research 2017, 6, 874. [Google Scholar] [CrossRef] [Green Version]
- Loboda, A.; Sobczak, M.; Jozkowicz, A.; Dulak, J. TGF-β1/Smads and miR-21 in renal fibrosis and inflammation. Mediat. Inflamm. 2016, 2016, 8319283. [Google Scholar] [CrossRef]
- Farris, A.B.; Colvin, R.B. Renal interstitial fibrosis: Mechanisms and evaluation in current opinion in nephrology and hypertension. Curr. Opin. Nephrol. Hypertens. 2013, 21, 289–300. [Google Scholar] [CrossRef]
- Avihingsanon, Y.; Benjachat, T.; Tassanarong, A.; Sodsai, P.; Kittikovit, V.; Hirankarn, N. Decreased renal expression of vascular endothelial growth factor in lupus nephritis is associated with worse prognosis. Kidney Int. 2009, 75, 1340–1348. [Google Scholar] [CrossRef] [Green Version]
- Kassimatis, T.I.; Nomikos, A.; Giannopoulou, I.; Lymperopoulos, A.; Moutzouris, D.A.; Varakis, I.; Nakopoulou, L. Transcription factor Sp1 expression is upregulated in human glomerulonephritis: Correlation with pSmad2/3 and p300 expression and renal injury. Ren. Fail. 2010, 32, 243–253. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, Y.; Xiong, M.; Fang, L.; Wen, P.; Cao, H.; Yang, J.; Dai, C.; He, W. Sp1 mediates microRNA-29c-regulated type I collagen production in renal tubular epithelial cells. Exp. Cell Res. 2013, 319, 2254–2265. [Google Scholar] [CrossRef]
- Gu, L.; Ni, Z.; Qian, J.; Tomino, Y. Pravastatin inhibits carboxymethyllysine-induced monocyte chemoattractant protein 1 expression in podocytes via prevention of signalling events. Nephron Exp. Nephrol. 2007, 106, e1–e10. [Google Scholar] [CrossRef]

| Characteristics | Lupus Nephritis (n = 45) | Healthy Control (n = 20) | ||
|---|---|---|---|---|
| Low CI (N = 18) | Moderate CI (N = 21) | High CI (N = 6) | ||
| Demographic | ||||
| Age, year | 33 ± 7 | 29 ± 5 | 30 ± 4 | 28 ± 5 |
| Sex, male/female | 7/11 | 8/13 | 3/3 | 8/12 |
| Race/ethnicity, n (%) | ||||
| Caucasian | 18 | 20 | 6 | 19 |
| Hispanic | 0 | 1 | 0 | 1 |
| Laboratory Parameters | ||||
| Serum creatinine, mg/Dl | 0.9 ± 0.3 | 1.1 ± 0.8 | 1.1 ± 0.6 | 0.7 ± 0.2 |
| eGFR (mL/min) | 92.3 ± 26.7 | 78.6 ± 29.3 | 78.8 ± 42.6 | 97.2 ± 31.7 |
| BUN (mmol/l) | 4.7 ± 1.2 | 4.6 ± 2.9 | 4.3 ± 1.7 | 5.0 ± 1.5 |
| Anti-dsDNA Abs, IU/mL | 333 ± 104 | 343 ± 107 | 357 ± 143 | n.d. |
| Serum C3, mg/dL | 69.5 ± 16.1 | 87.5 ± 12.1 | 74.5 ± 16.1 | n.d. |
| Serum C4, mg/dL | 8.3 ± 2.1 | 10.4 ± 3.5 | 11.2 ± 2.7 | n.d. |
| Proteinuria, g/24 h | 4.2 ± 2.4 | 3.6 ± 2.5 | 3.9 ± 4.7 | n.d. |
| Disease Index (SLEDAI-2K) | ||||
| Total SLEDAI score | 18 ± 2 | 14 ± 2 | 16 ± 3 | n.d. |
| Renal Biopsy, n (%) | ||||
| Class, n (%) | ||||
| III | 4 | 6 | 0 | n.d. |
| IV | 14 | 12 | 4 | n.d. |
| V | 0 | 3 | 1 | n.d. |
| Activity Index | 6.3 ± 3.7 | 5.3 ± 4.1 | 6.3 ± 3.2 | n.d. |
| Chronicity Index | 0.6 ± 0.4 | 3.5 ± 0.5 | 5.8 ± 1.2 | n.d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solé, C.; Moliné, T.; Vidal, M.; Ordi-Ros, J.; Cortés-Hernández, J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells 2019, 8, 773. https://doi.org/10.3390/cells8080773
Solé C, Moliné T, Vidal M, Ordi-Ros J, Cortés-Hernández J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells. 2019; 8(8):773. https://doi.org/10.3390/cells8080773
Chicago/Turabian StyleSolé, Cristina, Teresa Moliné, Marta Vidal, Josep Ordi-Ros, and Josefina Cortés-Hernández. 2019. "An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis" Cells 8, no. 8: 773. https://doi.org/10.3390/cells8080773