Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes
Abstract
:1. Introduction
2. Central Apparatus
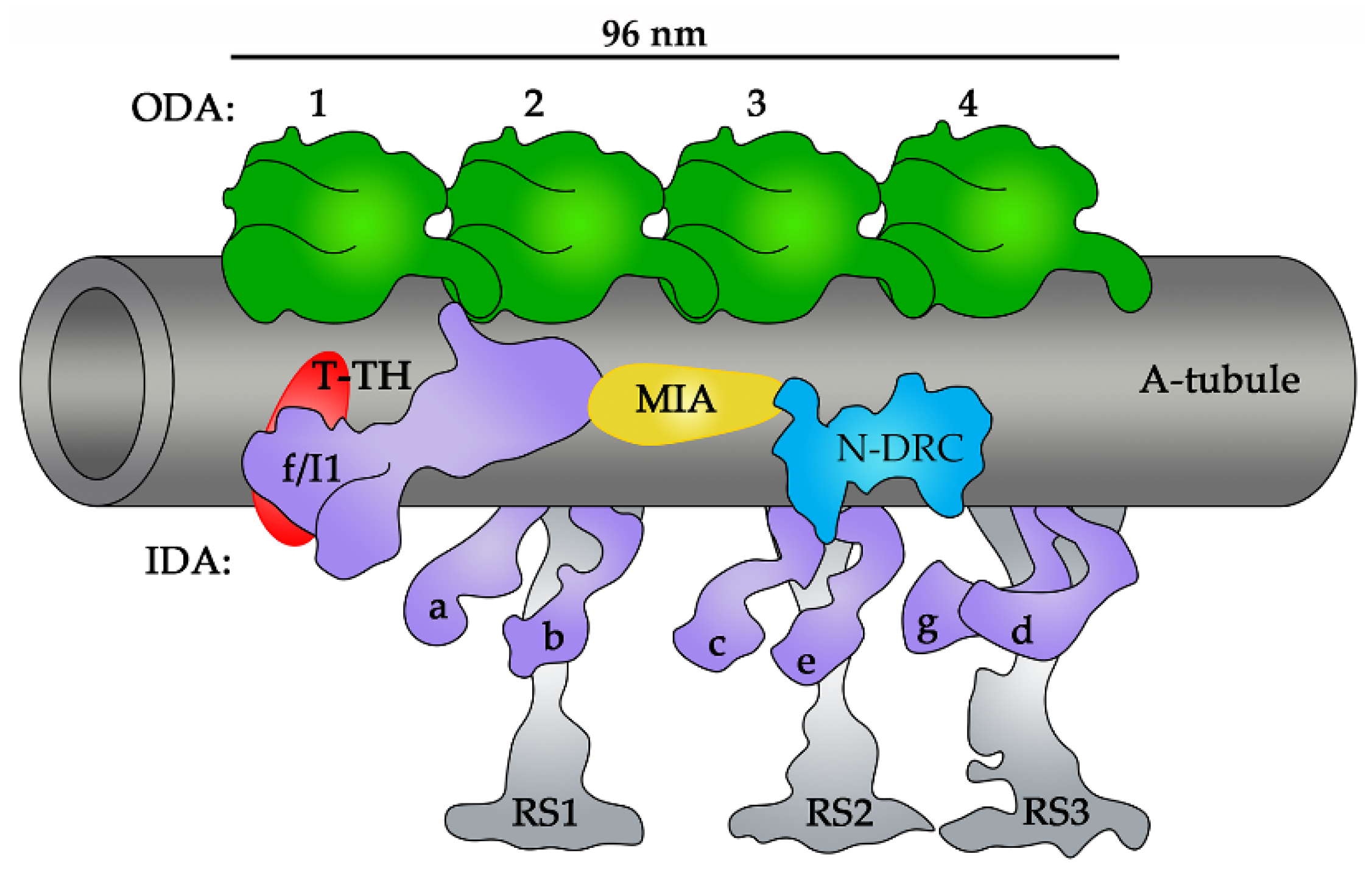
3. Outer Doublets and Microtubule Inner Proteins
4. Ciliary Ruler
5. Outer and Inner Dynein Arms
6. Nexin–Dynein Regulatory Complex
7. Radial Spokes
8. Small Complexes and Links
9. From Basic Science to Human Health
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garcia-Gonzalo, F.R.; Reiter, J.F. Open Sesame: How Transition Fibers and the Transition Zone Control Ciliary Composition. Cold Spring Harb. Perspect. Biol. 2017, 9, a028134. [Google Scholar] [CrossRef]
- Shahid, U.; Singh, P. Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs. Cells 2018, 7, 152. [Google Scholar] [CrossRef]
- Soares, H.; Carmona, B.; Nolasco, S.; Viseu Melo, L.; Gonçalves, J. Cilia Distal Domain: Diversity in Evolutionarily Conserved Structures. Cells 2019, 8, 160. [Google Scholar] [CrossRef]
- Nicastro, D.; Schwartz, C.; Pierson, J.; Gaudette, R.; Porter, M.E.; McIntosh, J.R. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 2006, 313, 944–948. [Google Scholar] [CrossRef]
- Carbajal-González, B.I.; Heuser, T.; Fu, X.; Lin, J.; Smith, B.W.; Mitchell, D.R.; Nicastro, D. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton 2013, 70, 101–120. [Google Scholar] [CrossRef]
- Li, J.B.; Gerdes, J.M.; Haycraft, C.J.; Fan, Y.; Teslovich, T.M.; May-Simera, H.; Li, H.; Blacque, O.E.; Li, L.; Leitch, C.C.; et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 2004, 117, 541–552. [Google Scholar] [CrossRef]
- Pazour, G.J.; Agrin, N.; Leszyk, J.; Witman, G.B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005, 170, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, K.; Bustamante-Marin, X.; Yin, W.; Goshe, M.B.; Ostrowski, L.E. Quantitative Proteomic Analysis of Human Airway Cilia Identifies Previously Uncharacterized Proteins of High Abundance. J. Proteome Res. 2017, 16, 1579–1592. [Google Scholar] [CrossRef] [Green Version]
- Wargo, M.J.; Smith, E.F. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 2003, 100, 137–142. [Google Scholar] [CrossRef]
- DiPetrillo, C.G.; Smith, E.F. The Pcdp1 complex coordinates the activity of dynein isoforms to produce wild-type ciliary motility. Mol. Biol. Cell 2011, 22, 4527–4538. [Google Scholar] [CrossRef]
- Zhao, L.; Hou, Y.; Picariello, T.; Craige, B.; Witman, G.B. Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 2019, 218, 2051–2070. [Google Scholar] [CrossRef] [Green Version]
- Adams, G.M.; Huang, B.; Piperno, G.; Luck, D.J. Central-pair microtubular complex of Chlamydomonas flagella: Polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 1981, 91, 69–76. [Google Scholar] [CrossRef]
- Teves, M.E.; Nagarkatti-Gude, D.R.; Zhang, Z.; Strauss, J.F., 3rd. Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton 2016, 73, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.F.; Lefebvre, P.A. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol. Biol. Cell 1997, 8, 455–467. [Google Scholar] [CrossRef]
- Zhang, Z.; Kostetskii, I.; Tang, W.; Haig-Ladewig, L.; Sapiro, R.; Wei, Z.; Patel, A.M.; Bennett, J.; Gerton, G.L.; Moss, S.B.; et al. Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol. Reprod. 2006, 74, 751–759. [Google Scholar] [CrossRef]
- Branche, C.; Kohl, L.; Toutirais, G.; Buisson, J.; Cosson, J.; Bastin, P. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 2006, 119, 3443–3455. [Google Scholar] [CrossRef] [Green Version]
- Portman, N.; Gull, K. The paraflagellar rod of kinetoplastid parasites: From structure to components and function. Int. J. Parasitol. 2010, 40, 135–148. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Li, Z.; Ping, P.; Wang, G.; Yuan, X.; Sun, F. Outer dense fibers stabilize the axoneme to maintain sperm motility. J. Cell. Mol. Med. 2018, 22, 1755–1768. [Google Scholar] [CrossRef]
- Rupp, G.; O’Toole, E.; Porter, M.E. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 2001, 12, 739–751. [Google Scholar] [CrossRef]
- Wargo, M.J.; Dymek, E.E.; Smith, E.F. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 2005, 118, 4655–4665. [Google Scholar] [CrossRef] [Green Version]
- Goduti, D.J.; Smith, E.F. Analyses of functional domains within the PF6 protein of the central apparatus reveal a role for PF6 sub-complex members in regulating flagellar beat frequency. Cytoskeleton 2012, 69, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Teves, M.E.; Zhang, Z.; Costanzo, R.M.; Henderson, S.C.; Corwin, F.D.; Zweit, J.; Sundaresan, G.; Subler, M.; Salloum, F.N.; Rubin, B.K.; et al. Sperm-associated antigen-17 gene is essential for motile cilia function and neonatal survival. Am. J. Respir. Cell Mol. Biol. 2013, 48, 765–772. [Google Scholar] [CrossRef]
- Mitchell, D.R.; Sale, W.S. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 1999, 144, 293–304. [Google Scholar] [CrossRef]
- Sironen, A.; Kotaja, N.; Mulhern, H.; Wyatt, T.A.; Sisson, J.H.; Pavlik, J.A.; Miiluniemi, M.; Fleming, M.D.; Lee, L. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol Reprod. 2011, 85, 690–701. [Google Scholar] [CrossRef]
- Zhang, H.; Mitchell, D.R. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J. Cell Sci. 2004, 117, 4179–4188. [Google Scholar] [CrossRef]
- Mitchell, B.F.; Pedersen, L.B.; Feely, M.; Rosenbaum, J.L.; Mitchell, D.R. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 2005, 16, 4509–4518. [Google Scholar] [CrossRef]
- DiPetrillo, C.G.; Smith, E.F. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J. Cell Biol. 2010, 189, 601–612. [Google Scholar] [CrossRef]
- Brown, J.M.; Dipetrillo, C.G.; Smith, E.F.; Witman, G.B. A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 2012, 125, 3904–3913. [Google Scholar] [CrossRef] [Green Version]
- Lechtreck, K.F.; Witman, G.B. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 2007, 176, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Lechtreck, K.F.; Delmotte, P.; Robinson, M.L.; Sanderson, M.J.; Witman, G.B. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 2008, 180, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, M.; Beech, P.L.; Katz, S.G.; Rosenbaum, J.L. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 1994, 125, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; O’toole, E.; Ghosh, S.; Mitchell, D.R. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 2004, 101, 17398–17403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.F.; Lefebvre, P.A. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 1996, 132, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Sapiro, R.; Kostetskii, I.; Olds-Clarke, P.; Gerton, G.L.; Radice, G.L.; Strauss, J.F., III. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 2002, 22, 6298–6305. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liu, H.; Zhu, L.; Chen, Y.; Zhao, H.; Zhang, W.; Li, F.; Xie, L.; Yan, X.; Zhu, X. Microtubule-bundling protein Spef1 enables mammalian ciliary central apparatus formation. J. Mol. Cell Biol. 2019, 11, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.G.; Sarafdar, R.B.; Chowdhury, T.S.; Sivadas, P.; Yang, P.; Dongre, P.M.; D’Souza, J.S. Myc-binding protein orthologue interacts with AKAP240 in the central pair apparatus of the Chlamydomonas flagella. BMC Cell Biol. 2016, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, Y.I.; Yao, E.; Lin, C.; Yang, J.H.; Wilson, C.W.; Gacayan, R.; Chuang, P.T. Fused (Stk36) is a ciliary protein required for central pair assembly and motile cilia orientation in the mammalian oviduct. Dev. Dyn. 2013, 242, 1307–1319. [Google Scholar] [CrossRef]
- Edelbusch, C.; Cindrić, S.; Dougherty, G.W.; Loges, N.T.; Olbrich, H.; Rivlin, J.; Wallmeier, J.; Pennekamp, P.; Amirav, I.; Omran, H. Mutation of serine/threonine protein kinase 36 (STK36) causes primary ciliary dyskinesia with a central pair defect. Hum. Mutat. 2017, 38, 964–969. [Google Scholar] [CrossRef]
- Dymek, E.E.; Lefebvre, P.A.; Smith, E.F. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot. Cell 2004, 3, 870–879. [Google Scholar] [CrossRef]
- Dymek, E.E.; Smith, E.F. PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J. Cell Sci. 2012, 125, 3357–3366. [Google Scholar] [CrossRef]
- Sharma, N.; Bryant, J.; Wloga, D.; Donaldson, R.; Davis, R.C.; Jerka-Dziadosz, M.; Gaertig, J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J. Cell Biol. 2007, 178, 1065–1079. [Google Scholar] [CrossRef] [Green Version]
- Nicastro, D.; Fu, X.; Heuser, T.; Tso, A.; Porter, M.E.; Linck, R.W. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. USA 2011, 108, E845–E853. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, A.; Obbineni, J.M.; Bui, K.H.; Shibata, K.; Toyoshima, Y.Y.; Ishikawa, T. α- and β-Tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo-electron microscopy and tomography. Structure 2015, 23, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Liu, D.; Kastritis, P.L.; Basu, K.; Hsu, T.C.; Yang, S.; Bui, K.H. Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat. Commun. 2017, 8, 15035. [Google Scholar] [CrossRef] [PubMed]
- Pigino, G.; Maheshwari, A.; Bui, K.H.; Shingyoji, C.; Kamimura, S.; Ishikawa, T. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J. Struct. Biol. 2012, 178, 199–206. [Google Scholar] [CrossRef]
- Yanagisawa, H.A.; Mathis, G.; Oda, T.; Hirono, M.; Richey, E.A.; Ishikawa, H.; Marshall, W.F.; Kikkawa, M.; Qin, H. FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. Mol. Biol. Cell 2014, 25, 1472–1483. [Google Scholar] [CrossRef] [PubMed]
- Dymek, E.E.; Lin, J.; Fu, G.; Porter, M.; Nicastro, D.; Smith, E.F. PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. Mol. Biol. Cell 2019, mbcE19010063. [Google Scholar] [CrossRef]
- Laligné, C.; Klotz, C.; de Loubresse, N.G.; Lemullois, M.; Hori, M.; Laurent, F.X.; Papon, J.F.; Louis, B.; Cohen, J.; Koll, F. Bug22p, a conserved centrosomal/ciliary protein also present in higher plants, is required for an effective ciliary stroke in Paramecium. Eukaryot. Cell 2010, 9, 645–655. [Google Scholar] [CrossRef]
- Sui, H.; Downing, K.H. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature 2006, 442, 475–478. [Google Scholar] [CrossRef] [Green Version]
- Ichikawa, M.; Bui, K.H. Microtubule inner proteins: A meshwork of luminal proteins stabilizing the doublet microtubule. Bioessays 2018, 40. [Google Scholar] [CrossRef]
- Stoddard, D.; Zhao, Y.; Bayless, B.A.; Gui, L.; Louka, P.; Dave, D.; Suryawanshi, S.; Tomasi, R.F.-X.; Dupuis-Williams, P.; Baroud, C.N.; et al. Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol. Biol. Cell 2018, 29, 2566–2577. [Google Scholar] [CrossRef] [PubMed]
- Owa, M.; Uchihashi, T.; Yanagisawa, H.A.; Yamano, T.; Iguchi, H.; Fukuzawa, H.; Wakabayashi, K.I.; Ando, T.; Kikkawa, M. Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat. Commun. 2019, 10, 1143. [Google Scholar] [CrossRef] [PubMed]
- Linck, R.; Fu, X.; Lin, J.; Ouch, C.; Schefter, A.; Steffen, W.; Warren, P.; Nicastro, D. Insights into the structure and function of ciliary and flagellar doublet microtubules tektins, Ca2+-binding proteins, and stable protofilaments. J. Biol. Chem. 2014, 289, 17427–17444. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Brown, J.A.; Yagi, T.; Norrander, J.M.; Hirono, M.; Eccleston, E.; Kamiya, R.; Linck, R.W. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J. Biol. Chem. 2003, 278, 7725–7734. [Google Scholar] [CrossRef] [PubMed]
- Kilburn, C.L.; Pearson, C.G.; Romijn, E.P.; Meehl, J.B.; Giddings, T.H., Jr.; Culver, B.P.; Yates, J.R., 3rd; Winey, M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007, 178, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirima, J.; Oiwa, K. Flagellar-associated protein FAP85 is a microtubule inner protein that stabilizes microtubules. Cell Struct. Funct. 2018, 43, 1–14. [Google Scholar] [CrossRef]
- Oda, T.; Yanagisawa, H.; Kamiya, R.; Kikkawa, M. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 2014, 346, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Becker-Heck, A.; Zohn, I.E.; Okabe, N.; Pollock, A.; Lenhart, K.B.; Sullivan-Brown, J.; McSheene, J.; Loges, N.T.; Olbrich, H.; Haeffner, K.; et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 2011, 43, 79–84. [Google Scholar] [CrossRef]
- Merveille, A.C.; Davis, E.E.; Becker-Heck, A.; Legendre, M.; Amirav, I.; Bataille, G.; Belmont, J.; Beydon, N.; Billen, F.; Clément, A.; et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011, 43, 72–78. [Google Scholar] [CrossRef]
- Antony, D.; Becker-Heck, A.; Zariwala, M.A.; Schmidts, M.; Onoufriadis, A.; Forouhan, M.; Wilson, R.; Taylor-Cox, T.; Dewar, A.; Jackson, C.; et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat. 2013, 34, 462–472. [Google Scholar] [CrossRef]
- Owa, M.; Furuta, A.; Usukura, J.; Arisaka, F.; King, S.M.; Witman, G.B.; Kamiya, R.; Wakabayashi, K. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc. Natl. Acad. Sci. USA 2014, 111, 9461–9466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koutoulis, A.; Pazour, G.J.; Wilkerson, C.G.; Inaba, K.; Sheng, H.; Takada, S.; Witman, G.B. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 1997, 137, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Mosley, M.; Montes-Berrueta, D.; Hou, Y.; Yang, F.; Scarbrough, C.; Witman, G.B.; Wirschell, M. Characterization of a new oda3 allele, oda3-6, defective in assembly of the outer dynein arm-docking complex in Chlamydomonas reinhardtii. PLoS ONE 2017, 12, e0173842. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Wilkerson, C.G.; Wakabayashi, K.; Kamiya, R.; Witman, G.B. The outer dynein arm-docking complex: Composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell 2002, 13, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.M.; Inaba, K.; Pazour, G.J.; Takada, S.; Wakabayashi, K.; Wilkerson, C.G.; Kamiya, R.; Witman, G.B. DC3, the 21-kDa subunit of the outer dynein arm docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol. Biol. Cell 2003, 14, 3650–3663. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.M.; Yagi, T.; Kamiya, R.; Witman, G.B. DC3, the smallest subunit of the Chlamydomonas flagellar outer dynein arm-docking complex, is a redox-sensitive calcium-binding protein. J. Biol. Chem. 2003, 278, 42652–42659. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Takada, S.; Witman, G.B.; Kamiya, R. Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell. Motil. Cytoskelet. 2001, 48, 277–286. [Google Scholar] [CrossRef]
- Haimo, L.T.; Fenton, R.D. Microtubule crossbridging by chlamydomonas dynein. Cell Motil. 1984, 4, 371–385. [Google Scholar] [CrossRef]
- Oda, T.; Abe, T.; Yanagisawa, H.; Kikkawa, M. Docking-complex-independent alignment of Chlamydomonas outer dynein arms with 24-nm periodicity in vitro. J. Cell Sci. 2016, 8, 1547–1551. [Google Scholar] [CrossRef]
- King, S.M.; Patel-King, R.S. The oligomeric outer dynein arm assembly factor CCDC103 is tightly integrated within the ciliary axoneme and exhibits periodic binding to microtubules. J. Biol. Chem. 2015, 290, 7388–7401. [Google Scholar] [CrossRef]
- Knowles, M.R.; Leigh, M.W.; Ostrowski, L.E.; Huang, L.; Carson, J.L.; Hazucha, M.J.; Yin, W.; Berg, J.S.; Davis, S.D.; Dell, S.D.; et al. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 92, 99–106. [Google Scholar] [CrossRef]
- Onoufriadis, A.; Paff, T.; Antony, D.; Shoemark, A.; Micha, D.; Kuyt, B.; Schmidts, M.; Petridi, S.; Dankert-Roelse, J.E.; Haarman, E.G.; et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 92, 88–98. [Google Scholar] [CrossRef]
- Hjeij, R.; Lindstrand, A.; Francis, R.; Zariwala, M.A.; Liu, X.; Li, Y.; Damerla, R.; Dougherty, G.W.; Abouhamed, M.; Olbrich, H.; et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013, 93, 357–367. [Google Scholar] [CrossRef]
- Hjeij, R.; Onoufriadis, A.; Watson, C.M.; Slagle, C.E.; Klena, N.T.; Dougherty, G.W.; Kurkowiak, M.; Loges, N.T.; Diggle, C.P.; Morante, N.F.; et al. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014, 95, 257–274. [Google Scholar] [CrossRef]
- Wallmeier, J.; Shiratori, H.; Dougherty, G.W.; Edelbusch, C.; Hjeij, R.; Loges, N.T.; Menchen, T.; Olbrich, H.; Pennekamp, P.; Raidt, J.; et al. TTC25 Deficiency Results in Defects of the Outer Dynein Arm Docking Machinery and Primary Ciliary Dyskinesia with Left-Right Body Asymmetry Randomization. Am. J. Hum. Genet. 2016, 99, 460–469. [Google Scholar] [CrossRef]
- Dean, A.B.; Mitchell, D.R. Late steps in cytoplasmic maturation of assembly-competent axonemal outer arm dynein in Chlamydomonas require interaction of ODA5 and ODA10 in a complex. Mol. Biol. Cell 2015, 26, 3596–3605. [Google Scholar] [CrossRef]
- King, S.M. Axonemal Dynein Arms. Cold Spring Harb. Perspect. Biol. 2016, 8, a028100. [Google Scholar] [CrossRef]
- Bui, K.H.; Sakakibara, H.; Movassagh, T.; Oiwa, K.; Ishikawa, T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 2008, 183, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Huizar, R.L.; Lee, C.; Boulgakov, A.A.; Horani, A.; Tu, F.; Marcotte, E.M.; Brody, S.L.; Wallingford, J.B. A liquid-like organelle at the root of motile ciliopathy. eLife 2018, 7, e38497. [Google Scholar] [CrossRef]
- Duquesnoy, P.; Escudier, E.; Vincensini, L.; Freshour, J.; Bridoux, A.M.; Coste, A.; Deschildre, A.; de Blic, J.; Legendre, M.; Montantin, G.; et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009, 85, 890–896. [Google Scholar] [CrossRef]
- Loges, N.T.; Olbrich, H.; Becker-Heck, A.; Häffner, K.; Heer, A.; Reinhard, C.; Schmidts, M.; Kispert, A.; Zariwala, M.A.; Leigh, M.W.; et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009, 85, 883–889. [Google Scholar] [CrossRef]
- Hartill, V.L.; van de Hoek, G.; Patel, M.P.; Little, R.; Watson, C.M.; Berry, I.R.; Shoemark, A.; Abdelmottaleb, D.; Parkes, E.; Bacchelli, C.; et al. DNAAF1 links heart laterality with the AAA+ ATPase RUVBL1 and ciliary intraflagellar transport. Hum. Mol. Genet. 2018, 27, 529–545. [Google Scholar] [CrossRef]
- Omran, H.; Kobayashi, D.; Olbrich, H.; Tsukahara, T.; Loges, N.T.; Hagiwara, H.; Zhang, Q.; Leblond, G.; O’Toole, E.; Hara, C.; et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 2008, 456, 611–616. [Google Scholar] [CrossRef] [Green Version]
- Mitchison, H.M.; Schmidts, M.; Loges, N.T.; Freshour, J.; Dritsoula, A.; Hirst, R.A.; O’Callaghan, C.; Blau, H.; Al Dabbagh, M.; Olbrich, H.; et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012, 44, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, G.; Vesterlund, L.; Hultenby, K.; Tapia-Pa’ez, I.; Kere, J. The zebrafish orthologue of the dyslexia candidate gene DYX1C1 is essential for cilia growth and function. PLoS ONE 2013, 8, e63123. [Google Scholar] [CrossRef]
- Tarkar, A.; Loges, N.T.; Slagle, C.E.; Francis, R.; Dougherty, G.W.; Tamayo, J.V.; Shook, B.; Cantino, M.; Schwartz, D.; Jahnke, C.; et al. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013, 45, 995–1003. [Google Scholar] [CrossRef]
- Yamamoto, R.; Obbineni, J.M.; Alford, L.M.; Ide, T.; Owa, M.; Hwang, J.; Kon, T.; Inaba, K.; James, N.; King, S.M.; et al. Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. PLoS Genet. 2017, 13, e1006996. [Google Scholar] [CrossRef]
- Horani, A.; Druley, T.E.; Zariwala, M.A.; Patel, A.C.; Levinson, B.T.; Van Arendonk, L.G.; Thornton, K.C.; Giacalone, J.C.; Albee, A.J.; Wilson, K.S.; et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012, 91, 685–693. [Google Scholar] [CrossRef]
- Diggle, C.P.; Moore, D.J.; Mali, G.; zur Lage, P.; Ait-Lounis, A.; Schmidts, M.; Shoemark, A.; Garcia Munoz, A.; Halachev, M.R.; Gautier, P.; et al. HEATR2 plays a conserved role in assembly of the ciliary motile apparatus. PLoS Genet. 2014, 10, e1004577. [Google Scholar] [CrossRef]
- Olcese, C.; Patel, M.P.; Shoemark, A.; Kiviluoto, S.; Legendre, M.; Williams, H.J.; Vaughan, C.K.; Hayward, J.; Goldenberg, A.; Emes, R.D.; et al. X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 2017, 8, 14279. [Google Scholar] [CrossRef] [Green Version]
- Paff, T.; Loges, N.T.; Aprea, I.; Wu, K.; Bakey, Z.; Haarman, E.G.; Daniels, J.M.A.; Sistermans, E.A.; Bogunovic, N.; Dougherty, G.W.; et al. Mutations in PIH1D3 Cause X-Linked Primary Ciliary Dyskinesia with Outer and Inner Dynein Arm Defects. Am. J. Hum. Genet. 2017, 100, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Höben, I.M.; Hjeij, R.; Olbrich, H.; Dougherty, G.W.; Nöthe-Menchen, T.; Aprea, I.; Frank, D.; Pennekamp, P.; Dworniczak, B.; Wallmeier, J.; et al. Mutations in C11orf70 Cause Primary Ciliary Dyskinesia with Randomization of Left/Right Body Asymmetry Due to Defects of Outer and Inner Dynein Arms. Am. J. Hum. Genet. 2018, 102, 973–984. [Google Scholar] [CrossRef] [Green Version]
- Fassad, M.R.; Shoemark, A.; le Borgne, P.; Koll, F.; Patel, M.; Dixon, M.; Hayward, J.; Richardson, C.; Frost, E.; Jenkins, L.; et al. C11orf70 Mutations Disrupting the Intraflagellar Transport-Dependent Assembly of Multiple Axonemal Dyneins Cause Primary Ciliary Dyskinesia. Am. J. Hum. Genet. 2018, 102, 956–972. [Google Scholar] [CrossRef] [Green Version]
- Kott, E.; Duquesnoy, P.; Copin, B.; Legendre, M.; Dastot-Le Moal, F.; Montantin, G.; Jeanson, L.; Tamalet, A.; Papon, J.F.; Siffroi, J.P.; et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012, 91, 958–964. [Google Scholar] [CrossRef]
- Horani, A.; Ferkol, T.W.; Shoseyov, D.; Wasserman, M.G.; Oren, Y.S.; Kerem, B.; Amirav, I.; Cohen-Cymberknoh, M.; Dutcher, S.K.; Brody, S.L.; et al. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS ONE 2013, 8, e59436. [Google Scholar] [CrossRef]
- Inaba, Y.; Shinohara, K.; Botilde, Y.; Nabeshima, R.; Takaoka, K.; Ajima, R.; Lamri, L.; Takeda, H.; Saga, Y.; Nakamura, T.; et al. Transport of the outer dynein arm complex to cilia requires a cytoplasmic protein Lrrc6. Genes Cells 2016, 21, 728–739. [Google Scholar] [CrossRef] [Green Version]
- Moore, D.J.; Onoufriadis, A.; Shoemark, A.; Simpson, M.A.; zur Lage, P.I.; de Castro, S.C.; Bartoloni, L.; Gallone, G.; Petridi, S.; Woollard, W.J.; et al. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013, 93, 346–356. [Google Scholar] [CrossRef]
- Zariwala, M.A.; Gee, H.Y.; Kurkowiak, M.; Al-Mutairi, D.A.; Leigh, M.W.; Hurd, T.W.; Hjeij, R.; Dell, S.D.; Chaki, M.; Dougherty, G.W.; et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013, 93, 336–345. [Google Scholar] [CrossRef]
- Mali, G.R.; Yeyati, P.L.; Mizuno, S.; Dodd, D.O.; Tennant, P.A.; Keighren, M.A.; Zur Lage, P.; Shoemark, A.; Garcia-Munoz, A.; Shimada, A.; et al. ZMYND10 functions in a chaperone relay during axonemal dynein assembly. eLife 2018, 7, e34389. [Google Scholar] [CrossRef]
- Cho, K.J.; Noh, S.H.; Han, S.M.; Choi, W.I.; Kim, H.Y.; Yu, S.; Lee, J.S.; Rim, J.H.; Lee, M.G.; Hildebrandt, F.; et al. ZMYND10 stabilizes intermediate chain proteins in the cytoplasmic pre-assembly of dynein arms. PLoS Genet. 2018, 14, e1007316. [Google Scholar] [CrossRef]
- Jaffe, K.M.; Grimes, D.T.; Schottenfeld-Roames, J.; Werner, M.E.; Ku, T.S.; Kim, S.K.; Pelliccia, J.L.; Morante, N.F.; Mitchell, B.J.; Burdine, R.D. c21orf59/kurly Controls Both Cilia Motility and Polarization. Cell Rep. 2016, 14, 1841–1849. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Wang, L.; Pan, J. Chlamydomonas WDR92 in association with R2TP-like complex and multiple DNAAFs to regulate ciliary dynein preassembly. J. Mol. Cell Biol. 2018, 1–11. [Google Scholar] [CrossRef]
- Zur Lage, P.; Stefanopoulou, P.; Styczynska-Soczka, K.; Quinn, N.; Mali, G.; von Kriegsheim, A.; Mill, P.; Jarman, A.P. Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J. Cell Biol. 2018, 217, 2583–2598. [Google Scholar] [CrossRef] [Green Version]
- Patel-King, R.S.; Sakato-Antoku, M.; Yankova, M.; King, S.M. WDR92 Is Required for Axonemal Dynein Heavy Chain Stability in Cytoplasm. Mol. Biol. Cell 2019, 30, 1781–1877. [Google Scholar] [CrossRef]
- Knowles, M.R.; Ostrowski, L.E.; Loges, N.T.; Hurd, T.; Leigh, M.W.; Huang, L.; Wolf, W.E.; Carson, J.L.; Hazucha, M.J.; Yin, W.; et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 2013, 93, 711–720. [Google Scholar] [CrossRef]
- Ahmed, N.T.; Gao, C.; Lucker, B.F.; Cole, D.G.; Mitchell, D.R. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J. Cell Biol. 2008, 183, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Taschner, M.; Mourão, A.; Awasthi, M.; Basquin, J.; Lorentzen, E. Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J. Biol. Chem. 2017, 292, 7462–7473. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Witman, G.B. The N-terminus of IFT46 mediates intraflagellar transport of outer arm dynein and its cargo-adaptor ODA16. Mol. Biol. Cell 2017, 28, 2420–2433. [Google Scholar] [CrossRef]
- Shamoto, N.; Narita, K.; Kubo, T.; Oda, T.; Takeda, S. CFAP70 Is a Novel Axoneme-Binding Protein That Localizes at the Base of the Outer Dynein Arm and Regulates Ciliary Motility. Cells 2018, 7, 124. [Google Scholar] [CrossRef]
- Satir, P.; Heuser, T.; Sale, W.S. A Structural Basis for How Motile Cilia Beat. Bioscience 2014, 64, 1073–1083. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Nicastro, D. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 2018, 360, eaar1968. [Google Scholar] [CrossRef]
- Heuser, T.; Raytchev, M.; Krell, J.; Porter, M.E.; Nicastro, D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 2009, 187, 921–933. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, I.R. Cilia and flagella of eukaryotes. J. Cell Biol. 1981, 91, 107–124. [Google Scholar] [CrossRef]
- Summers, K.E.; Gibbons, I.R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA 1971, 68, 3092–3096. [Google Scholar] [CrossRef]
- Warner, F.D. Ciliary inter-microtubule bridges. J. Cell Sci. 1976, 20, 101–114. [Google Scholar]
- Bui, K.H.; Yagi, T.; Yamamoto, R.; Kamiya, R.; Ishikawa, T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 2012, 198, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Oda, T.; Yagi, T.; Yanagisawa, H.; Kikkawa, M. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr. Biol. 2013, 23, 656–664. [Google Scholar] [CrossRef]
- Yamamoto, R.; Song, K.; Yanagisawa, H.A.; Fox, L.; Yagi, T.; Wirschell, M.; Hirono, M.; Kamiya, R.; Nicastro, D.; Sale, W.S. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J. Cell Biol. 2013, 201, 263–278. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Tritschler, D.; Song, K.; Barber, C.F.; Cobb, J.S.; Porter, M.E.; Nicastro, D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 2011, 286, 29175–29191. [Google Scholar] [CrossRef]
- Bower, R.; Tritschler, D.; Vanderwaal, K.; Perrone, C.A.; Mueller, J.; Fox, L.; Sale, W.S.; Porter, M.E. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell 2013, 24, 1134–1152. [Google Scholar] [CrossRef]
- Oda, T.; Yanagisawa, H.; Kikkawa, M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol. Biol. Cell 2015, 26, 294–304. [Google Scholar] [CrossRef]
- Song, K.; Awata, J.; Tritschler, D.; Bower, R.; Witman, G.B.; Porter, M.E.; Nicastro, D. In situ localization of N- and C-termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP-tag and cryo-electron tomography. J. Biol. Chem. 2015, 290, 5341–5353. [Google Scholar] [CrossRef]
- Bower, R.; Tritschler, D.; Mills, K.V.; Heuser, T.; Nicastro, D.; Porter, M.E. DRC2/CCDC65 is a central hub for assembly of the nexin-dynein regulatory complex and other regulators of ciliary and flagellar motility. Mol. Biol. Cell 2017, 29, 137–153. [Google Scholar] [CrossRef]
- Awata, J.; Song, K.; Lin, J.; King, S.M.; Sanderson, M.J.; Nicastro, D.; Witman, G.B. DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol. Biol. Cell 2015, 26, 2788–2800. [Google Scholar] [CrossRef] [Green Version]
- Austin-Tse, C.; Halbritter, J.; Zariwala, M.A.; Gilberti, R.M.; Gee, H.Y.; Hellman, N.; Pathak, N.; Liu, Y.; Panizzi, J.R.; Patel-King, R.S.; et al. Zebrafish Ciliopathy Screen Plus Human Mutational Analysis Identifies C21orf59 and CCDC65 Defects as Causing Primary Ciliary Dyskinesia. Am. J. Hum. Genet. 2013, 93, 672–686. [Google Scholar] [CrossRef] [Green Version]
- Horani, A.; Brody, S.L.; Ferkol, T.W.; Shoseyov, D.; Wasserman, M.G.; Ta-shma, A.; Wilson, K.S.; Bayly, P.V.; Amirav, I.; Cohen-Cymberknoh, M.; et al. CCDC65 mutation causes primary ciliary dyskinesia with normal ultrastructure and hyperkinetic cilia. PLoS ONE 2013, 26, e72299. [Google Scholar] [CrossRef]
- Wirschell, M.; Olbrich, H.; Werner, C.; Tritschler, D.; Bower, R.; Sale, W.S.; Loges, N.T.; Pennekamp, P.; Lindberg, S.; Stenram, U.; et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat. Genet. 2013, 45, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Olbrich, H.; Cremers, C.; Loges, N.T.; Werner, C.; Nielsen, K.G.; Marthin, J.K.; Philipsen, M.; Wallmeier, J.; Pennekamp, P.; Menchen, T.; et al. Loss-of-Function GAS8 Mutations Cause Primary Ciliary Dyskinesia and Disrupt the Nexin-Dynein Regulatory Complex. Am. J. Hum. Genet. 2015, 97, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Lewis, W.R.; Malarkey, E.B.; Tritschler, D.; Bower, R.; Pasek, R.C.; Porath, J.D.; Birket, S.E.; Saunier, S.; Antignac, C.; Knowles, M.R.; et al. Mutation of Growth Arrest Specific 8 Reveals a Role in Motile Cilia Function and Human Disease. PLoS Genet. 2016, 12, e1006220. [Google Scholar] [CrossRef]
- Kubo, T.; Yanagisawa, H.A.; Yagi, T.; Hirono, M.; Kamiya, R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 2010, 20, 441–445. [Google Scholar] [CrossRef]
- Suryavanshi, S.; Eddé, B.; Fox, L.A.; Guerrero, S.; Hard, R.; Hennessey, T.; Kabi, A.; Malison, D.; Pennock, D.; Sale, W.S.; et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 2010, 20, 435–440. [Google Scholar] [CrossRef]
- Kubo, T.; Oda, T. Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility. Mol. Biol. Cell 2017, 28, 2260–2266. [Google Scholar] [CrossRef]
- Alford, L.M.; Stoddard, D.; Li, J.H.; Hunter, E.L.; Tritschler, D.; Bower, R.; Nicastro, D.; Porter, M.E.; Sale, W.S. The nexin link and B-tubule glutamylation maintain the alignment of outer doublets in the ciliary axoneme. Cytoskeleton 2016, 73, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Pigino, G.; Ishikawa, T. Axonemal radial spokes: 3D structure, function and assembly. Bioarchitecture 2012, 2, 50–58. [Google Scholar] [CrossRef]
- Pigino, G.; Bui, K.H.; Maheshwari, A.; Lupetti, P.; Diener, D.; Ishikawa, T. Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 2011, 195, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Heuser, T.; Carbajal-Gonzalez, B.I.; Song, K.; Nicastro, D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton 2012, 69, 88–100. [Google Scholar] [CrossRef]
- Urbanska, P.; Song, K.; Joachimiak, E.; Krzemien-Ojak, L.; Koprowski, P.; Hennessey, T.; Jerka-Dziadosz, M.; Fabczak, H.; Gaertig, J.; Nicastro, D.; et al. The calmodulin- and spoke-associated complex proteins FAP61 and FAP251 build the radial spoke 3 base complex in cilia. Mol. Biol. Cell 2015, 26, 1463–1475. [Google Scholar] [CrossRef]
- Piperno, G.; Huang, B.; Ramanis, Z.; Luck, D.J. Radial spokes of Chlamydomonas flagella: Polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 1981, 88, 73–79. [Google Scholar] [CrossRef]
- Yang, P.; Diener, D.R.; Rosenbaum, J.L.; Sale, W.S. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 2001, 153, 1315–1326. [Google Scholar] [CrossRef]
- Yang, P.; Diener, D.R.; Yang, C.; Kohno, T.; Pazour, G.J.; Dienes, J.M.; Agrin, N.S.; King, S.M.; Sale, W.S.; Kamiya, R.; et al. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 2006, 119, 1165–1174. [Google Scholar] [CrossRef]
- Zhu, X.; Poghosyan, E.; Rezabkova, L.; Mehall, B.; Sakakibara, H.; Hirono, M.; Kamiya, R.; Ishikawa, T.; Yang, P. The roles of a flagellar HSP40 ensuring rhythmic beating. Mol. Biol. Cell 2019, 30, 228–241. [Google Scholar] [CrossRef]
- Vasudevan, K.K.; Song, K.; Alford, L.M.; Sale, W.S.; Dymek, E.E.; Smith, E.F.; Hennessey, T.; Joachimiak, E.; Urbanska, P.; Wloga, D.; et al. FAP206 is a microtubule-docking adapter for ciliary radial spoke 2 and dynein c. Mol. Biol. Cell 2015, 26, 696–710. [Google Scholar] [CrossRef]
- Dymek, E.E.; Smith, E.F. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 2007, 179, 515–526. [Google Scholar] [CrossRef]
- Dymek, E.E.; Heuser, T.; Nicastro, D.; Smith, E.F. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol. Biol. Cell 2011, 22, 2520–2531. [Google Scholar] [CrossRef]
- Heuser, T.; Dymek, E.E.; Lin, J.; Smith, E.F.; Nicastro, D. The CSC connects three major axonemal complexes involved in dynein regulation. Mol. Biol. Cell 2012, 23, 3143–3155. [Google Scholar] [CrossRef]
- Satouh, Y.; Inaba, K. Proteomic characterization of sperm radial spokes identifies a novel spoke protein with an ubiquitin domain. FEBS Lett. 2009, 583, 2201–2207. [Google Scholar] [CrossRef] [Green Version]
- Gopal, R.; Foster, K.W.; Yang, P. The DPY-30 domain and its flanking sequence mediate the assembly and modulation of flagellar radial spoke complexes. Mol. Cell. Biol. 2012, 32, 4012–4024. [Google Scholar] [CrossRef]
- Sivadas, P.; Dienes, J.M.; St Maurice, M.; Meek, W.D.; Yang, P. A flagellar A-kinase anchoring protein with two amphipathic helices forms a structural scaffold in the radial spoke complex. J. Cell Biol. 2012, 199, 639–651. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, T. Cryo-electron tomography of motile cilia and flagella. Cilia 2015, 4, 3. [Google Scholar] [CrossRef]
- Kikushima, K. Central pair apparatus enhances outer-arm dynein activities through regulation of inner-arm dyneins. Cell Motil. Cytoskelet. 2009, 66, 272–280. [Google Scholar] [CrossRef]
- Heuser, T.; Barber, C.F.; Lin, J.; Krell, J.; Rebesco, M.; Porter, M.E.; Nicastro, D. Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc. Natl. Acad. Sci. USA 2012, 109, E2067–E2076. [Google Scholar] [CrossRef]
- Fu, G.; Wang, Q.; Phan, N.; Urbanska, P.; Joachimiak, E.; Lin, J.; Wloga, D.; Nicastro, D. The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol. Biol. Cell 2018, 29, 1048–1059. [Google Scholar] [CrossRef]
- Kubo, T.; Hou, Y.; Cochran, D.A.; Witman, G.B.; Oda, T. A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol. Biol. Cell 2018, 29, 1060–1074. [Google Scholar] [CrossRef]
- Urbanska, P.; Joachimiak, E.; Bazan, R.; Fu, G.; Poprzeczko, M.; Fabczak, H.; Nicastro, D.; Wloga, D. Ciliary proteins Fap43 and Fap44 interact with each other and are essential for proper cilia and flagella beating. Cell. Mol. Life Sci. 2018, 75, 4479–4493. [Google Scholar] [CrossRef] [Green Version]
- Horani, A.; Ferkol, T.W. Advances in the Genetics of Primary Ciliary Dyskinesia: Clinical Implications. Chest 2018, 154, 645–652. [Google Scholar] [CrossRef]
- Nsota Mbango, J.F.; Coutton, C.; Arnoult, C.; Ray, P.F.; Touré, A. Genetic causes of male infertility: Snapshot on morphological abnormalities of the sperm flagellum. Basic Clin. Androl. 2019, 29, 2. [Google Scholar] [CrossRef]
- Diener, D.R.; Lupetti, P.; Rosenbaum, J.L. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol. 2015, 25, 379–384. [Google Scholar] [CrossRef]
- Dean, S.; Moreira-Leite, F.; Varga, V.; Gull, K. Cilium transition zone proteome reveals compartmentalization and differential dynamics of ciliopathy complexes. Proc. Natl. Acad. Sci. USA 2016, 113, E5135–E5143. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, J.; Pelletier, L. The Ciliary Transition Zone: Finding the Pieces and Assembling the Gate. Mol. Cells 2017, 40, 243–253. [Google Scholar] [CrossRef] [Green Version]
- Croft, J.T.; Zabeo, D.; Subramanian, R.; Höög, J.L. Composition, structure and function of the eukaryotic flagellum distal tip. Essays Biochem. 2018, 62, 815–828. [Google Scholar] [CrossRef] [Green Version]
- Louka, P.; Vasudevan, K.K.; Guha, M.; Joachimiak, E.; Wloga, D.; Tomasi, R.F.; Baroud, C.N.; Dupuis-Williams, P.; Galati, D.F.; Pearson, C.G.; et al. Proteins that control the geometry of microtubules at the ends of cilia. J. Cell Biol. 2018, 217, 4298–4313. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, M.J.; Phetruen, T.; Fisher, R.L.; Chen, K.; Pentecost, B.T.; Gomez, G.; Ounjai, P.; Sui, H. The Developmental Process of the Growing Motile Ciliary Tip Region. Sci. Rep. 2018, 8, 7977. [Google Scholar] [CrossRef]
- Taschner, M.; Lorentzen, E. The Intraflagellar Transport Machinery. Cold Spring Harb. Perspect. Biol. 2016, 8, a028092. [Google Scholar] [CrossRef] [Green Version]
- Prevo, B.; Scholey, J.M.; Peterman, E.J.G. Intraflagellar transport: Mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 2017, 284, 2905–2931. [Google Scholar] [CrossRef]
- Wingfield, J.L.; Lechtreck, K.F.; Lorentzen, E. Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 2018, 62, 753–763. [Google Scholar] [CrossRef]
- Wirschell, M.; Yamamoto, R.; Alford, L.; Gokhale, A.; Gaillard, A.; Sale, W.S. Regulation of ciliary motility: Conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme. Arch. Biochem. Biophys. 2011, 510, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Wloga, D.; Joachimiak, E.; Louka, P.; Gaertig, J. Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harb. Perspect. Biol. 2017, 9, a028159. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osinka, A.; Poprzeczko, M.; Zielinska, M.M.; Fabczak, H.; Joachimiak, E.; Wloga, D. Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes. Cells 2019, 8, 730. https://doi.org/10.3390/cells8070730
Osinka A, Poprzeczko M, Zielinska MM, Fabczak H, Joachimiak E, Wloga D. Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes. Cells. 2019; 8(7):730. https://doi.org/10.3390/cells8070730
Chicago/Turabian StyleOsinka, Anna, Martyna Poprzeczko, Magdalena M. Zielinska, Hanna Fabczak, Ewa Joachimiak, and Dorota Wloga. 2019. "Ciliary Proteins: Filling the Gaps. Recent Advances in Deciphering the Protein Composition of Motile Ciliary Complexes" Cells 8, no. 7: 730. https://doi.org/10.3390/cells8070730




