Ameliorating the Fibrotic Remodeling of the Heart through Direct Cardiac Reprogramming
Abstract
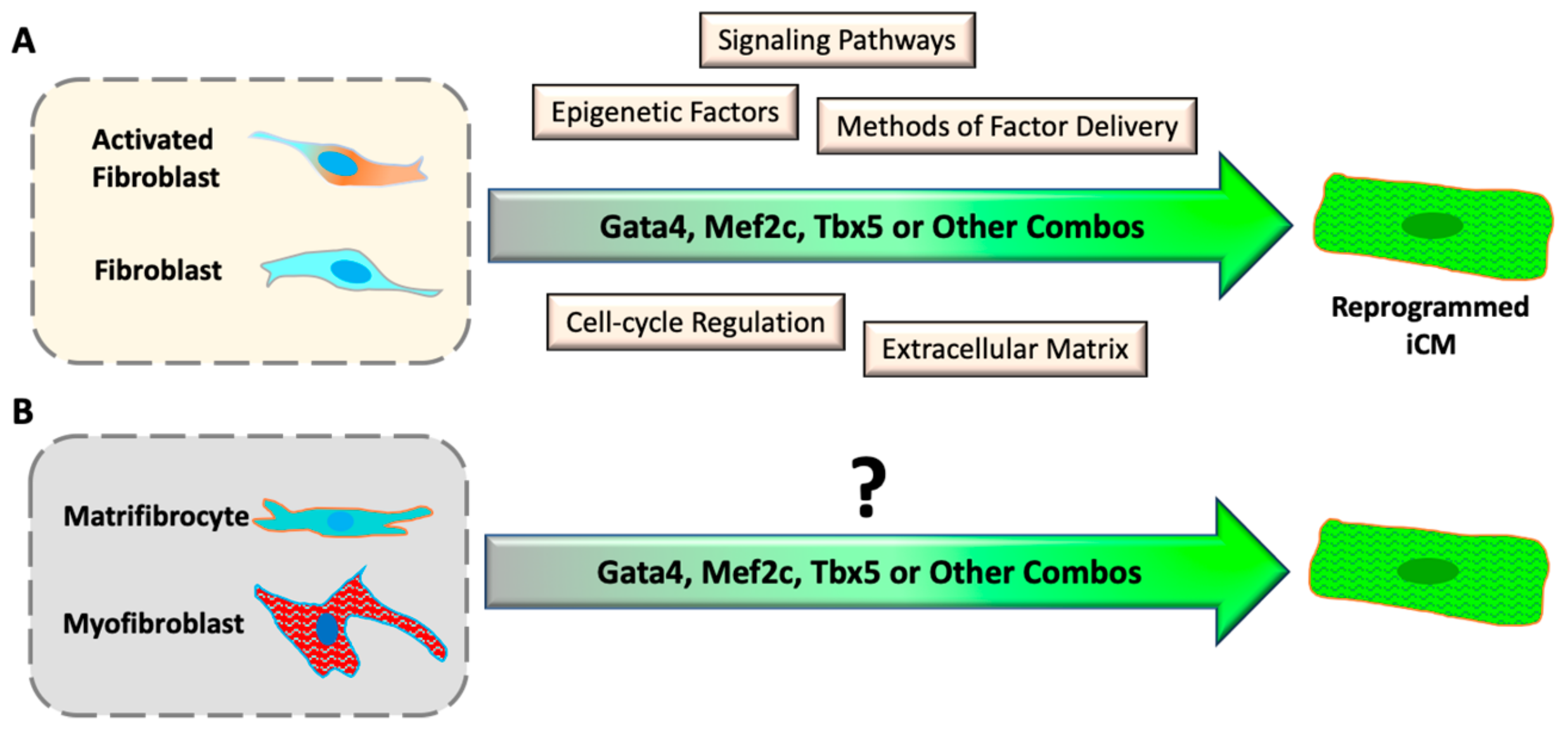
:1. Introduction
2. Pathological Remodeling of the Heart
3. Development of Direct Cardiac Reprogramming
3.1. Direct Reprogramming of Mouse Fibroblasts into iCMs In Vitro
3.2. In Situ Reprogramming of iCMs in The Heart
3.3. Direct Cardiac Reprogramming of Human Fibroblasts
3.4. Direct Reprogramming to Multipotent Cardiac Progenitors
4. Mechanistical Understanding of Direct Cardiac Reprogramming
4.1. Activation of Signaling Pathways During Reprogramming
4.2. Epigenetic Barriers of Reprogramming
4.3. Cell-Cycle Regulation During Direct Cardiac Reprogramming
4.4. Modification of Extracellular Matrix
4.5. Non-Genomic Integration Methods for Direct Cardiac Reprogramming
5. Perspective to Translate Direct Cardiac Reprogramming
5.1. Understanding the Mystery of In Vivo Reprogramming
5.2. Converting Chronic Fibrotic Scar into Myocardium
5.3. Direct Cardiac Reprogramming of Human Cells
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Benjamin, E.J.; Blaha, M.J.; Chiuve, S.E.; Cushman, M.; Das, S.R.; Deo, R.; de Ferranti, S.D.; Floyd, J.; Fornage, M.; Gillespie, C.; et al. Heart Disease and Stroke Statistics-2017 Update: A Report from the American Heart Association. Circulation 2017, 135, e146–e603. [Google Scholar] [CrossRef] [PubMed]
- Savi, M.; Bocchi, L.; Fiumana, E.; Karam, J.P.; Frati, C.; Bonafe, F.; Cavalli, S.; Morselli, P.G.; Guarnieri, C.; Caldarera, C.M.; et al. Enhanced engraftment and repairing ability of human adipose-derived stem cells, conveyed by pharmacologically active microcarriers continuously releasing HGF and IGF-1, in healing myocardial infarction in rats. J. Biomed. Mater. Res. Part A 2015, 103, 3012–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, G.; Iyer, V.; Lee, T.C.; Canty, J.M., Jr. Autologous mesenchymal stem cells mobilize cKit+ and CD133+ bone marrow progenitor cells and improve regional function in hibernating myocardium. Circ. Res. 2011, 109, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chiu, S.; Liang, X.; Gao, F.; Zhang, Z.; Liao, S.; Liang, Y.; Chai, Y.H.; Low, D.J.; Tse, H.F.; et al. Rap1-mediated nuclear factor-kappaB (NF-kappaB) activity regulates the paracrine capacity of mesenchymal stem cells in heart repair following infarction. Cell Death Discov. 2015, 1, 15007. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, X.; Zhang, Y.; Liang, X.; Ding, Y.; Xu, Y.; Fang, Z.; Zhang, F. Enhanced cell survival and paracrine effects of mesenchymal stem cells overexpressing hepatocyte growth factor promote cardioprotection in myocardial infarction. Exp. Cell Res. 2016, 344, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, E.; Mummery, C.L.; Bellin, M. Human heart disease: Lessons from human pluripotent stem cell-derived cardiomyocytes. Cell. Mol. Life Sci. 2017, 74, 3711–3739. [Google Scholar] [CrossRef] [PubMed]
- Mandel, Y.; Weissman, A.; Schick, R.; Barad, L.; Novak, A.; Meiry, G.; Goldberg, S.; Lorber, A.; Rosen, M.R.; Itskovitz-Eldor, J.; et al. Human embryonic and induced pluripotent stem cell-derived cardiomyocytes exhibit beat rate variability and power-law behavior. Circulation 2012, 125, 883–893. [Google Scholar] [CrossRef]
- Moreau, A.; Boutjdir, M.; Chahine, M. Induced pluripotent stem-cell-derived cardiomyocytes: Cardiac applications, opportunities, and challenges. Can. J. Physiol. Pharmacol. 2017, 95, 1108–1116. [Google Scholar] [CrossRef]
- Smart, N.; Bollini, S.; Dube, K.N.; Vieira, J.M.; Zhou, B.; Davidson, S.; Yellon, D.; Riegler, J.; Price, A.N.; Lythgoe, M.F.; et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011, 474, 640. [Google Scholar] [CrossRef]
- Ellison, G.M.; Torella, D.; Dellegrottaglie, S.; Perez-Martinez, C.; Perez de Prado, A.; Vicinanza, C.; Purushothaman, S.; Galuppo, V.; Iaconetti, C.; Waring, C.D.; et al. Endogenous cardiac stem cell activation by insulin-like growth factor-1/hepatocyte growth factor intracoronary injection fosters survival and regeneration of the infarcted pig heart. J. Am. Coll. Cardiol. 2011, 58, 977–986. [Google Scholar] [CrossRef]
- Urbanek, K.; Rota, M.; Cascapera, S.; Bearzi, C.; Nascimbene, A.; De Angelis, A.; Hosoda, T.; Chimenti, S.; Baker, M.; Limana, F.; et al. Cardiac stem cells possess growth factor-receptor systems that after activation regenerate the infarcted myocardium, improving ventricular function and long-term survival. Circ. Res. 2005, 97, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Linke, A.; Muller, P.; Nurzynska, D.; Casarsa, C.; Torella, D.; Nascimbene, A.; Castaldo, C.; Cascapera, S.; Bohm, M.; Quaini, F.; et al. Stem cells in the dog heart are self-renewing, clonogenic, and multipotent and regenerate infarcted myocardium, improving cardiac function. Proc. Natl. Acad. Sci. USA 2005, 102, 8966–8971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campa, V.M.; Gutierrez-Lanza, R.; Cerignoli, F.; Diaz-Trelles, R.; Nelson, B.; Tsuji, T.; Barcova, M.; Jiang, W.; Mercola, M. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J. Cell Biol. 2008, 183, 129–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, F.B.; Hauck, L.; Cardoso, M.C.; Leonhardt, H.; Dietz, R.; von Harsdorf, R. A mammalian myocardial cell-free system to study cell cycle reentry in terminally differentiated cardiomyocytes. Circ. Res. 1999, 85, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A.; Obana, M.; Mitsuhara, Y.; Orimoto, A.; Nakayasu, Y.; Yamashita, T.; Fukada, S.I.; Maeda, M.; Nakayama, H.; Fujio, Y. Adult murine cardiomyocytes exhibit regenerative activity with cell cycle reentry through STAT3 in the healing process of myocarditis. Sci. Rep. 2017, 7, 1407. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.M.A.; Ang, Y.S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018, 173, 104–116. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Inagawa, K.; Miyamoto, K.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Umei, T.; Wada, R.; Katsumata, Y.; Kaneda, R.; Nakade, K.; et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 1147–1156. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.-d.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzahor, E.; Poss, K.D. Cardiac regeneration strategies: Staying young at heart. Science 2017, 356, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Pu, W.T. Strategies for cardiac regeneration and repair. Sci. Transl. Med. 2014, 6, 239rv1. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Sadahiro, T.; Ieda, M. Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. Int. J. Mol. Sci. 2018, 19, 2629. [Google Scholar] [CrossRef] [PubMed]
- Vaseghi, H.; Liu, J.; Qian, L. Molecular barriers to direct cardiac reprogramming. Protein Cell 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Z.; Zhao, Z.-A.A.; Shen, Z. Direct reprogramming of fibroblasts into cardiomyocytes. Stem Cell Res. Ther. 2017, 8, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frangogiannis, N.G. Pathophysiology of Myocardial Infarction. Compr. Physiol. 2015, 5, 1841–1875. [Google Scholar] [CrossRef]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef]
- Banerjee, I.; Fuseler, J.W.; Price, R.L.; Borg, T.K.; Baudino, T.A. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H1883–H1891. [Google Scholar] [CrossRef] [Green Version]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’Antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef] [Green Version]
- Souders, C.A.; Bowers, S.L.; Baudino, T.A. Cardiac fibroblast: The renaissance cell. Circ. Res. 2009, 105, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, E.M.; Kalluri, R. Origins of cardiac fibroblasts. Circ. Res. 2010, 107, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Dewald, O.; Zymek, P.; Winkelmann, K.; Koerting, A.; Ren, G.; Abou-Khamis, T.; Michael, L.H.; Rollins, B.J.; Entman, M.L.; Frangogiannis, N.G. CCL2/Monocyte Chemoattractant Protein-1 regulates inflammatory responses critical to healing myocardial infarcts. Circ. Res. 2005, 96, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G.; Michael, L.H.; Entman, M.L. Myofibroblasts in reperfused myocardial infarcts express the embryonic form of smooth muscle myosin heavy chain (SMemb). Cardiovasc. Res. 2000, 48, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Ivey, M.J.; Kuwabara, J.T.; Pai, J.T.; Moore, R.E.; Sun, Z.; Tallquist, M.D. Resident fibroblast expansion during cardiac growth and remodeling. J. Mol. Cell. Cardiol. 2018, 114, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.V.; Fagard, R.H.; Lijnen, P.J. Stimulation of collagen production by transforming growth factor-beta1 during differentiation of cardiac fibroblasts to myofibroblasts. Hypertension 2002, 39, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Bujak, M.; Frunza, O.; Dobaczewski, M.; Gonzalez-Quesada, C.; Lu, B.; Gerard, C.; Frangogiannis, N.G. CXCR3-independent actions of the CXC chemokine CXCL10 in the infarcted myocardium and in isolated cardiac fibroblasts are mediated through proteoglycans. Cardiovasc. Res. 2014, 103, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Bujak, M.; Dobaczewski, M.; Gonzalez-Quesada, C.; Xia, Y.; Leucker, T.; Zymek, P.; Veeranna, V.; Tager, A.M.; Luster, A.D.; Frangogiannis, N.G. Induction of the CXC chemokine interferon-gamma-inducible protein 10 regulates the reparative response following myocardial infarction. Circ. Res. 2009, 105, 973–983. [Google Scholar] [CrossRef]
- Czubryt, M.P. Common threads in cardiac fibrosis, infarct scar formation, and wound healing. Fibrogenes. Tissue Repair 2012, 5, 19. [Google Scholar] [CrossRef]
- Desmouliere, A.; Redard, M.; Darby, I.; Gabbiani, G. Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar. Am. J. Pathol. 1995, 146, 56–66. [Google Scholar]
- Willems, I.E.; Havenith, M.G.; De Mey, J.G.; Daemen, M.J. The alpha-smooth muscle actin-positive cells in healing human myocardial scars. Am. J. Pathol. 1994, 145, 868–875. [Google Scholar] [PubMed]
- Fu, X.; Khalil, H.; Kanisicak, O.; Boyer, J.G.; Vagnozzi, R.J.; Maliken, B.D.; Sargent, M.A.; Prasad, V.; Valiente-Alandi, I.; Blaxall, B.C.; et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Investig. 2018, 128, 2127–2143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Costa, M.L.; Mermelstein, C.S.; Chagas, C.; Holtzer, S.; Holtzer, H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl. Acad. Sci. USA 1990, 87, 7988–7992. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Protze, S.; Khattak, S.; Poulet, C.; Lindemann, D.; Tanaka, E.M.; Ravens, U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 2012, 53, 323–332. [Google Scholar] [CrossRef]
- Hirai, H.; Katoku-Kikyo, N.; Keirstead, S.A.; Kikyo, N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc. Res. 2013, 100, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef]
- Stone, N.R.; Gifford, C.A.; Thomas, R.; Pratt, K.J.B.; Samse-Knapp, K.; Mohamed, T.M.A.; Radzinsky, E.M.; Schricker, A.; Yu, P.; Ivey, K.N.; et al. Unique Transcription Factor Functions Regulate Epigenetic and Transcriptional Dynamics During Cardiac Reprogramming. bioRxiv 2019. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, A.D.; Kim, L.J.; Nam, Y.J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019, 9, 6362. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Ieda, M.; Fu, J.; Qian, L. Cardiac repair with thymosin β4 and cardiac reprogramming factors. Ann. N. Y. Acad. Sci. 2012, 1270, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Finch, E.A.; Zhang, L.; Zhang, H.; Hodgkinson, C.P.; Pratt, R.E.; Rosenberg, P.B.; Mirotsou, M.; Dzau, V.J. MicroRNA induced cardiac reprogramming in vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015, 116, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, L.; Yin, C.; Liu, J.; Qian, L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc. Res. 2015, 108, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Welch, J.D.; Ma, H.; Zhou, Y.; Vaseghi, H.R.; Yu, S.; Wall, J.B.; Alimohamadi, S.; Zheng, M.; et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 2017. [Google Scholar] [CrossRef]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.-D.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef]
- Bektik, E.; Dennis, A.; Prasanna, P.; Madabhushi, A.; Fu, J.-D.D. Single cell qPCR reveals that additional HAND2 and microRNA-1 facilitate the early reprogramming progress of seven-factor-induced human myocytes. PLoS ONE 2017, 12, e0183000. [Google Scholar] [CrossRef]
- Mohamed, T.M.A.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.-S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming Clinical Perspective. Circulation 2016, 135, 978–995. [Google Scholar] [CrossRef]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalit, P.A.; Salick, M.R.; Nelson, D.O.; Squirrell, J.M.; Shafer, C.M.; Patel, N.G.; Saeed, I.; Schmuck, E.G.; Markandeya, Y.S.; Wong, R.; et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 2016, 18, 354–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Cao, N.; Huang, Y.; Spencer, I.C.; Fu, J.-d.; Yu, C.; Liu, K.; Nie, B.; Xu, T.; Li, K.; et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 2016, 18, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bektik, E.; Dennis, A.; Pawlowski, G.; Zhou, C.; Maleski, D.; Takahashi, S.; Laurita, K.R.; Deschênes, I.; Fu, J.-D.D. S-phase Synchronization Facilitates the Early Progression of Induced-Cardiomyocyte Reprogramming through Enhanced Cell-Cycle Exit. Int. J. Mol. Sci. 2018, 19, 1364. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.; Hashimoto, H.; Zhou, H.; Morales, M.G.; Chen, B.; Bassel-Duby, R.; Olson, E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017, 8, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 11864–11869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [Green Version]
- Yijing, G.; Ienglam, L.; Shuo, T.; Wenbin, G.; Karatas, H.; Yangbing, L.; Shaomeng, W.; Liu, L.; Zhong, W. Enhancing Cardiac Reprogramming by Suppressing Specific C-C Chemokine Signaling Pathways. bioRxiv 2019. [Google Scholar] [CrossRef]
- Muraoka, N.; Nara, K.; Tamura, F.; Kojima, H.; Yamakawa, H.; Sadahiro, T.; Miyamoto, K.; Isomi, M.; Haginiwa, S.; Tani, H.; et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 2019, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Chronis, C.; Fiziev, P.; Papp, B.; Butz, S.; Bonora, G.; Sabri, S.; Ernst, J.; Plath, K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 2017, 168, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.R.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 2013, 155, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Ang, C.E.; Davila, J.; Pak, C.; Mall, M.; Lee, Q.Y.; Ahlenius, H.; Jung, S.W.; Sudhof, T.C.; Wernig, M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014, 3, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Zheng, M.; Wang, L.; Zhou, Y.; Yin, C.; Liu, J.; Qian, L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016, 16, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, H.; Kikyo, N. Inhibitors of suppressive histone modification promote direct reprogramming of fibroblasts to cardiomyocyte-like cells. Cardiovasc. Res. 2014, 102, 188–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dal-Pra, S.; Hodgkinson, C.P.; Mirotsou, M.; Kirste, I.; Dzau, V.J. Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo. Circ. Res. 2017, 120, 1403–1413. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.-D.D.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Wang, Z.; Garry, G.A.; Malladi, V.S.; Botten, G.A.; Ye, W.; Zhou, H.; Osterwalder, M.; Dickel, D.E.; Visel, A.; et al. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell 2019. [Google Scholar] [CrossRef] [PubMed]
- Donaghey, J.; Thakurela, S.; Charlton, J.; Chen, J.S.; Smith, Z.D.; Gu, H.; Pop, R.; Clement, K.; Stamenova, E.K.; Karnik, R.; et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018, 50, 250–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.E.; Fraga, M.F.; Weaver, I.C.; Berdasco, M.; Szyf, M. Variations in DNA methylation patterns during the cell cycle of HeLa cells. Epigenetics 2007, 2, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Bou Kheir, T.; Lund, A.H. Epigenetic dynamics across the cell cycle. Essays Biochem. 2010, 48, 107–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wang, L.; Liu, Z.; Alimohamadi, S.; Yin, C.; Liu, J.; Qian, L. Comparative Gene Expression Analyses Reveal Distinct Molecular Signatures between Differentially Reprogrammed Cardiomyocytes. Cell Rep. 2017, 20, 3014–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Liu, Z.; Welch, J.D.; Gao, X.; Wang, L.; Garbutt, T.; Keepers, B.; Ma, H.; Prins, J.F.; Shen, W.; et al. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell 2019. [Google Scholar] [CrossRef]
- Takeuchi, T. Regulation of cardiomyocyte proliferation during development and regeneration. Dev. Growth Differ. 2014, 56, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Chopra, A.; Lin, V.; McCollough, A.; Atzet, S.; Prestwich, G.D.; Wechsler, A.S.; Murray, M.E.; Oake, S.A.; Kresh, J.Y.; Janmey, P.A. Reprogramming cardiomyocyte mechanosensing by crosstalk between integrins and hyaluronic acid receptors. J. Biomech. 2012, 45, 824–831. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.J.; Carag-Krieger, C.; Johnson, C.P.; Raab, M.; Tang, H.Y.; Speicher, D.W.; Sanger, J.W.; Sanger, J.M.; Discher, D.E. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: Scar-like rigidity inhibits beating. J. Cell Sci. 2008, 121, 3794–3802. [Google Scholar] [CrossRef]
- Ribeiro, A.J.; Ang, Y.S.; Fu, J.D.; Rivas, R.N.; Mohamed, T.M.; Higgs, G.C.; Srivastava, D.; Pruitt, B.L. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. USA 2015, 112, 12705–12710. [Google Scholar] [CrossRef] [Green Version]
- Sia, J.; Yu, P.; Srivastava, D.; Li, S. Effect of biophysical cues on reprogramming to cardiomyocytes. Biomaterials 2016, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Dal-Pra, S.; Mirotsou, M.; Jayawardena, T.M.; Hodgkinson, C.P.; Bursac, N.; Dzau, V.J. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, A.; Ma, H.; Yao, K.; Zhou, N.; Shen, L.; Zhang, C.; Zou, Y.; Ge, J. Infarcted myocardium-like stiffness contributes to endothelial progenitor lineage commitment of bone marrow mononuclear cells. J. Cell. Mol. Med. 2011, 15, 2245–2261. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, A.P.; DeLeon-Pennell, K.Y.; Ma, Y.; Halade, G.V.; Yabluchanskiy, A.; Iyer, R.P.; Flynn, E.; Cates, C.A.; Lindsey, M.L.; Han, H.C. Building a better infarct: Modulation of collagen cross-linking to increase infarct stiffness and reduce left ventricular dilation post-myocardial infarction. J. Mol. Cell. Cardiol. 2015, 85, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, K.; Takeuchi, Y. Gene therapy using retrovirus vectors: Vector development and biosafety at clinical trials. Uirusu 2015, 65, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Vrljicak, P.; Tao, S.; Varshney, G.K.; Quach, H.N.; Joshi, A.; LaFave, M.C.; Burgess, S.M.; Sampath, K. Genome-Wide Analysis of Transposon and Retroviral Insertions Reveals Preferential Integrations in Regions of DNA Flexibility. G3 2016, 6, 805–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathison, M.; Gersch, R.P.; Nasser, A.; Lilo, S.; Korman, M.; Fourman, M.; Hackett, N.; Shroyer, K.; Yang, J.; Ma, Y.; et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J. Am. Heart Assoc. 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Mathison, M.; Singh, V.P.; Chiuchiolo, M.J.; Sanagasetti, D.; Mao, Y.; Patel, V.B.; Yang, J.; Kaminsky, S.M.; Crystal, R.G.; Rosengart, T.K. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J. Thorac. Cardiovasc. Surg. 2017, 153, 329–339. [Google Scholar] [CrossRef]
- Nam, Y.-J.J.; Lubczyk, C.; Bhakta, M.; Zang, T.; Fernandez-Perez, A.; McAnally, J.; Bassel-Duby, R.; Olson, E.N.; Munshi, N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 2014, 141, 4267–4278. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.Y.; Jeong, S.-N.N.; Kang, J.-I.I.; Lee, S.-W.W. Chimeric Adeno-Associated Virus-Mediated Cardiovascular Reprogramming for Ischemic Heart Disease. ACS Omega 2018, 3, 5918–5925. [Google Scholar] [CrossRef]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Lee, E.; Kim, J.; Kwon, Y.-W.; Kwon, Y.; Kim, J. Efficient in vivo direct conversion of fibroblasts into cardiomyocytes using a nanoparticle-based gene carrier. Biomaterials 2018. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015, 25, 1013–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Tu, W.; Fu, Y.; Wang, J.; Xie, X. Chemical-induced cardiac reprogramming in vivo. Cell Res. 2018, 28, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.-D.D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, X.; Shen, Y.; Zhang, R.; Sugi, K.; Vasudevan, N.T.; Alaiti, M.A.; Sweet, D.R.; Zhou, L.; Qing, Y.; Gerson, S.L.; et al. Distinct roles of resident and nonresident macrophages in nonischemic cardiomyopathy. Proc. Natl. Acad. Sci. USA 2018, 115, E4661–E4669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golpanian, S.; Wolf, A.; Hatzistergos, K.E.; Hare, J.M. Rebuilding the Damaged Heart: Mesenchymal Stem Cells, Cell-Based Therapy, and Engineered Heart Tissue. Physiol. Rev. 2016, 96, 1127–1168. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Berry, E.C.; Fu, J.D.; Ieda, M.; Srivastava, D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat. Protoc. 2013, 8, 1204–1215. [Google Scholar] [CrossRef]
- Srivastava, D.; Ieda, M. Critical factors for cardiac reprogramming. Circ. Res. 2012, 111, 5–8. [Google Scholar] [CrossRef]
- Chen, J.X.; Krane, M.; Deutsch, M.A.; Wang, L.; Rav-Acha, M.; Gregoire, S.; Engels, M.C.; Rajarajan, K.; Karra, R.; Abel, E.D.; et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 50–55. [Google Scholar] [CrossRef]
- Cacchiarelli, D.; Qiu, X.; Srivatsan, S.; Manfredi, A.; Ziller, M.; Overbey, E.; Grimaldi, A.; Grimsby, J.; Pokharel, P.; Livak, K.J.; et al. Aligning Single-Cell Developmental and Reprogramming Trajectories Identifies Molecular Determinants of Myogenic Reprogramming Outcome. Cell Syst. 2018, 7, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.D.; Srivastava, D. Direct reprogramming of fibroblasts into cardiomyocytes for cardiac regenerative medicine. Circ. J. 2015, 79, 245–254. [Google Scholar] [CrossRef] [PubMed]


| Signaling Pathways | Mouse iCM Reprogramming | Human iCM Reprogramming |
|---|---|---|
| TGFβ Inhibition | Enhanced by suppression of Smad2 and 3 phosphorylation [64,65] | Enhanced [60] |
| Akt1 Activation | Enhanced through activation of mTORC1 and Foxo3a in embryonic but not in adult fibroblasts [68] | Unknown |
| JAK inhibition | Enhanced iCM quality [19] | Unknown |
| RhoA-ROCK Inhibition | Enhanced through suppression of SRF-signaling [65] | Unknown |
| Notch Inhibition | Enhanced through increased Mef2c activity [67] | Unknown |
| Wnt Inhibition | Enhanced through suppression of canonical Wnt [60] | Enhanced [60] |
| FFV (Fgf2, FGF10, VEGF) | Enhanced through activation of p38 MAPK and PI3K/Akt pathways [69] | Unknown |
| Inflammatory Signaling Pathways | ||
| Cyclooxygenase-2 Inhibition (by diclofenac) | Improved iCM quality through suppression of E2/PGE R4, cAMP/PKA, and IL1β [72] | Unknown |
| C-C chemokine inhibition | Enhanced by suppression of chemokine receptors [71] | Unknown |
| Transcriptional Regulators | ||
| ZNF281 Activation | Enhanced by cooperation with Gata4 and suppression of inflammatory response [70] | Unknown |
| ZFPM2 Activation | Unknown | Enhanced [58] |
| Epigenetic Factors | ||
| Bmi1 Inhibition | Improved induction of beating iCMs [80] | Unknown |
| Ezh2 Inhibition | Enhanced by suppression of H3K27me2 & H3K27me3 [78,79] | Unknown |
| G9a and GLP Inhibition | Enhanced by suppression of H3K9me & H3K9me2 [78] | Unknown |
| Topological Factors | ||
| Microgroove ECM | Improved through nuclear localization of Mkl1 [91] | Unknown |
| 3D Hydrogels | Improved by increased expression of MMPs [92] | Unknown |
| Cell-cycle Manipulations | ||
| S-phase Synchronization | Accelerated by enhancing cell-cycle exit [66] | Unknown |
| G2/M-phase Synchronization | Improved iCM quality and yield [56] | Unknown |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bektik, E.; Fu, J.-d. Ameliorating the Fibrotic Remodeling of the Heart through Direct Cardiac Reprogramming. Cells 2019, 8, 679. https://doi.org/10.3390/cells8070679
Bektik E, Fu J-d. Ameliorating the Fibrotic Remodeling of the Heart through Direct Cardiac Reprogramming. Cells. 2019; 8(7):679. https://doi.org/10.3390/cells8070679
Chicago/Turabian StyleBektik, Emre, and Ji-dong Fu. 2019. "Ameliorating the Fibrotic Remodeling of the Heart through Direct Cardiac Reprogramming" Cells 8, no. 7: 679. https://doi.org/10.3390/cells8070679





