CD44s Assembles Hyaluronan Coat on Filopodia and Extracellular Vesicles and Induces Tumorigenicity of MKN74 Gastric Carcinoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Creation of MKN74 Cell Lines and Cell Culture
2.2. Quantitative Real-Time RT-PCR (qRT-PCR)
2.3. Western Blotting
2.4. Nanoparticle Tracking Analysis
2.5. Nanoview Analysis
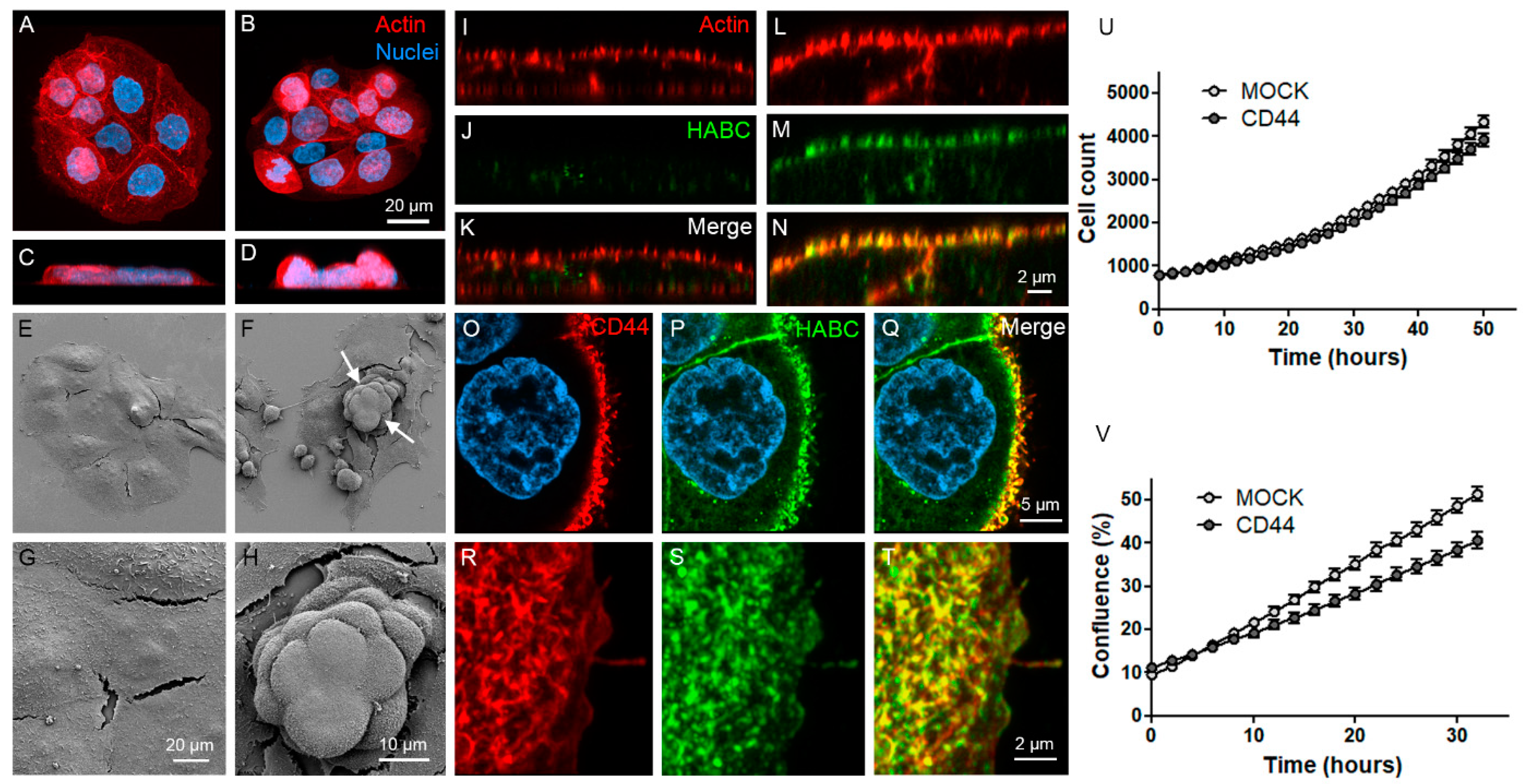
2.6. Proliferation Rate and Confluency
2.7. HA Assay
2.8. Immunostaining of CD44 and Staining of HA in Live and Fixed Cells
2.9. Vital Stainings
2.10. Confocal Imaging and Image Analysis
2.11. Immuno-Electron Microscopy of EVs
2.12. Scanning Electron Microcopy
2.13. Invasion and Spheroidal Assays
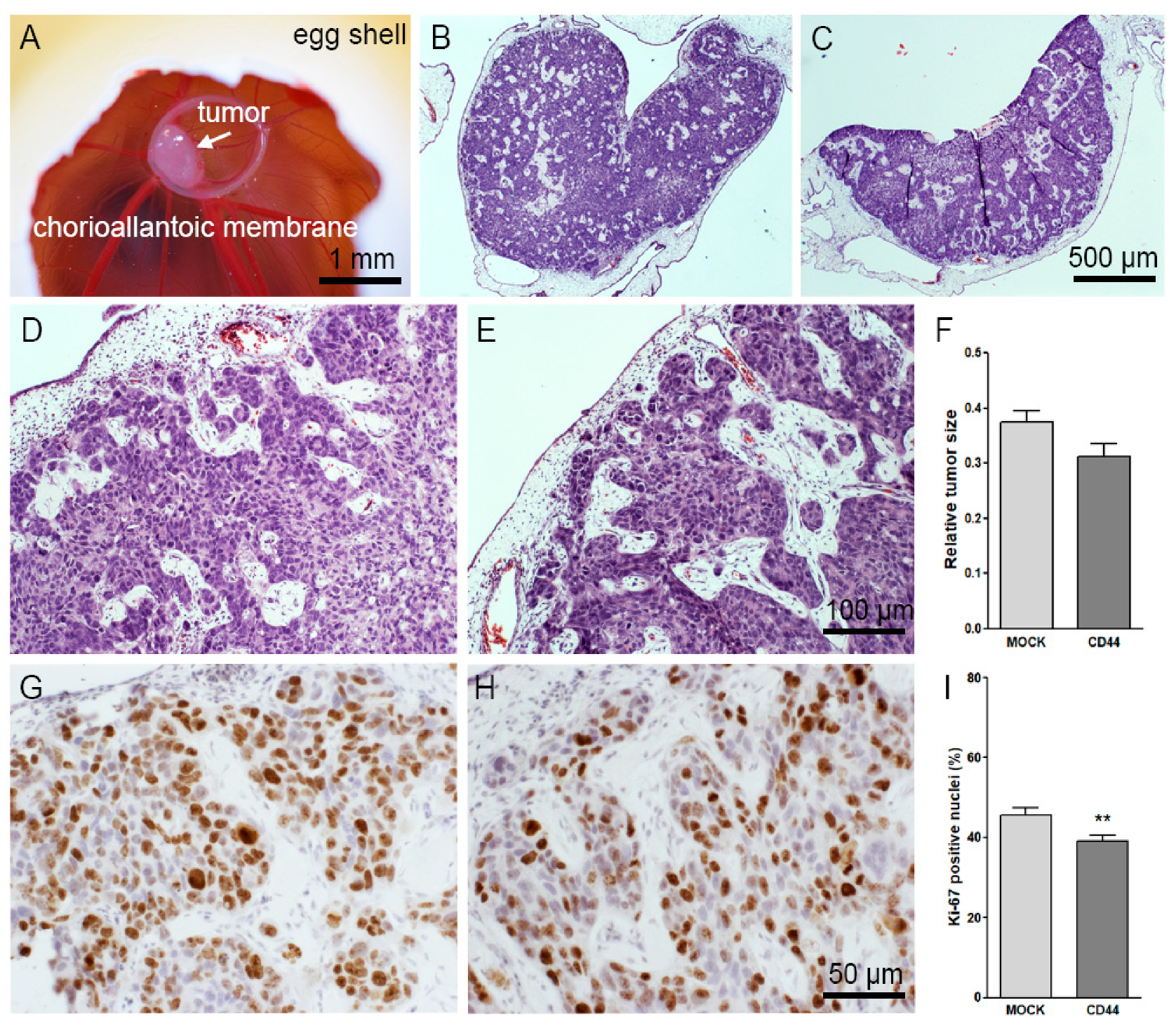
2.14. Chick Chorioallantoic Membrane (CAM) Assays
2.15. Staining and Analysis of CAM Tumor Sections
2.16. Statistical Methods
3. Results
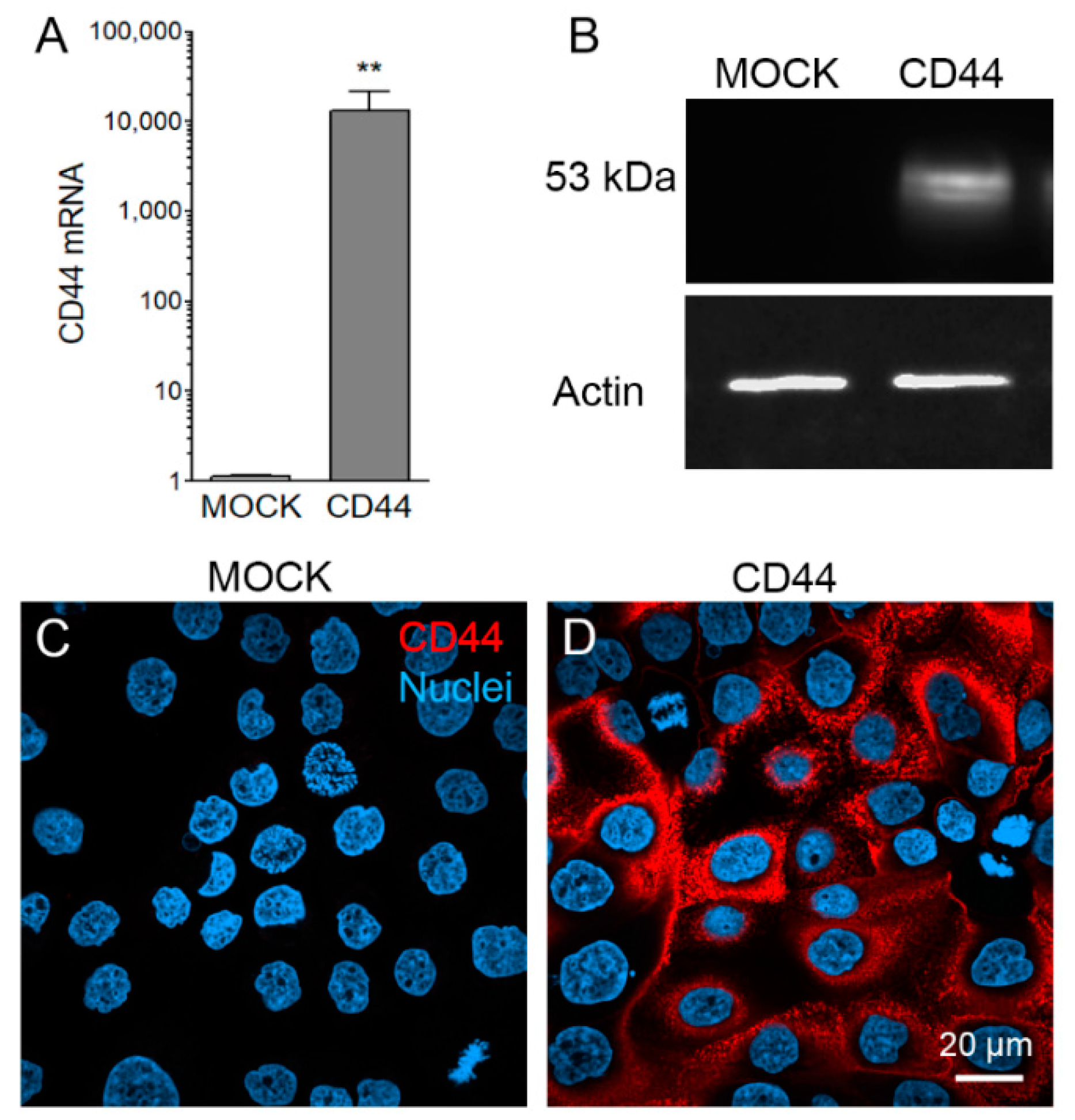
3.1. Stable CD44s Transfected MKN74 Cells Express High Levels of CD44s
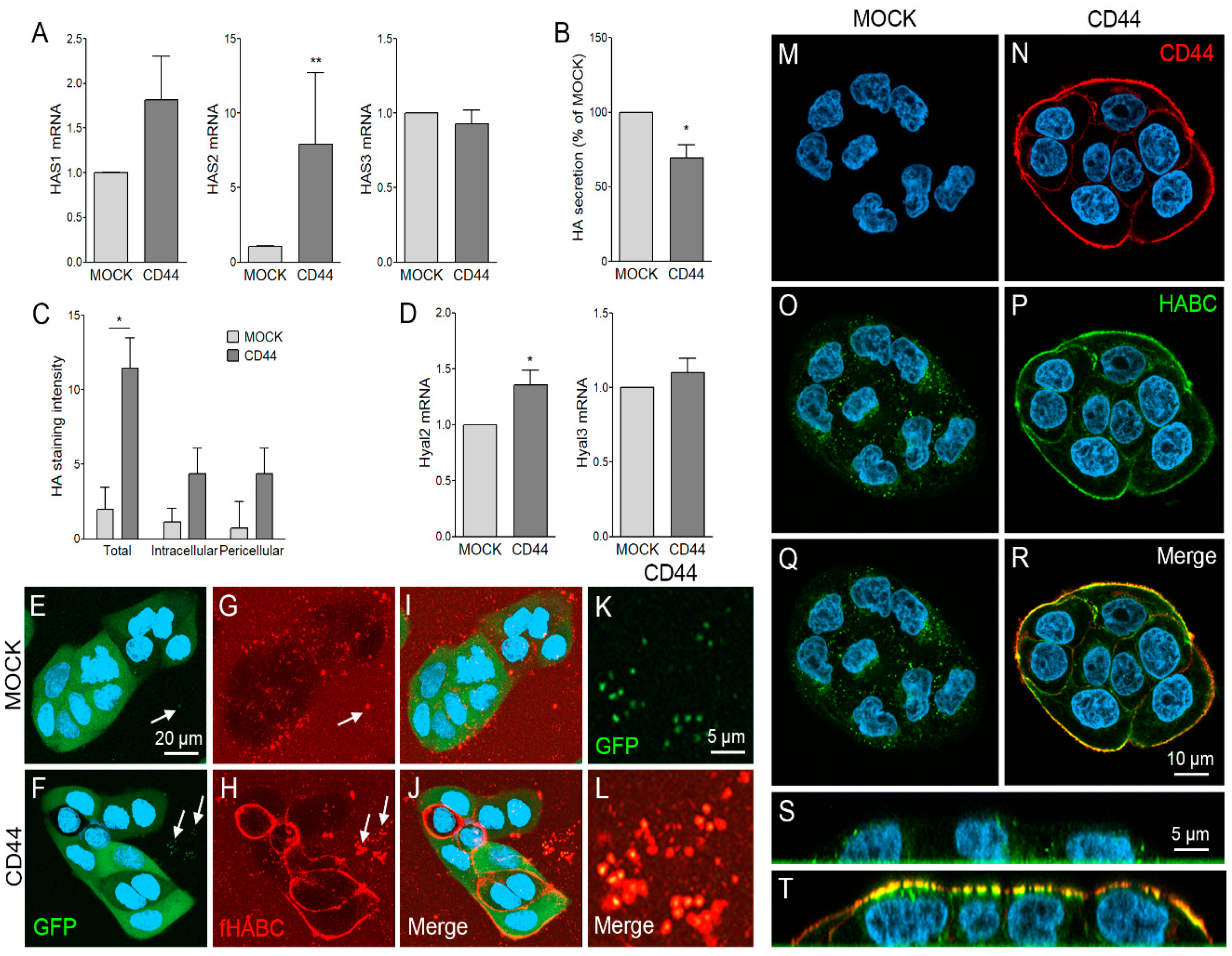
3.2. CD44s Regulates HA Metabolism of MKN74 Cell Lines
3.3. CD44s Accumulates on Filopodia and Binds HA on Filopodial Membranes
3.4. CD44s Is Accumulated on EVs but Does Not Affect EV Secretion Levels
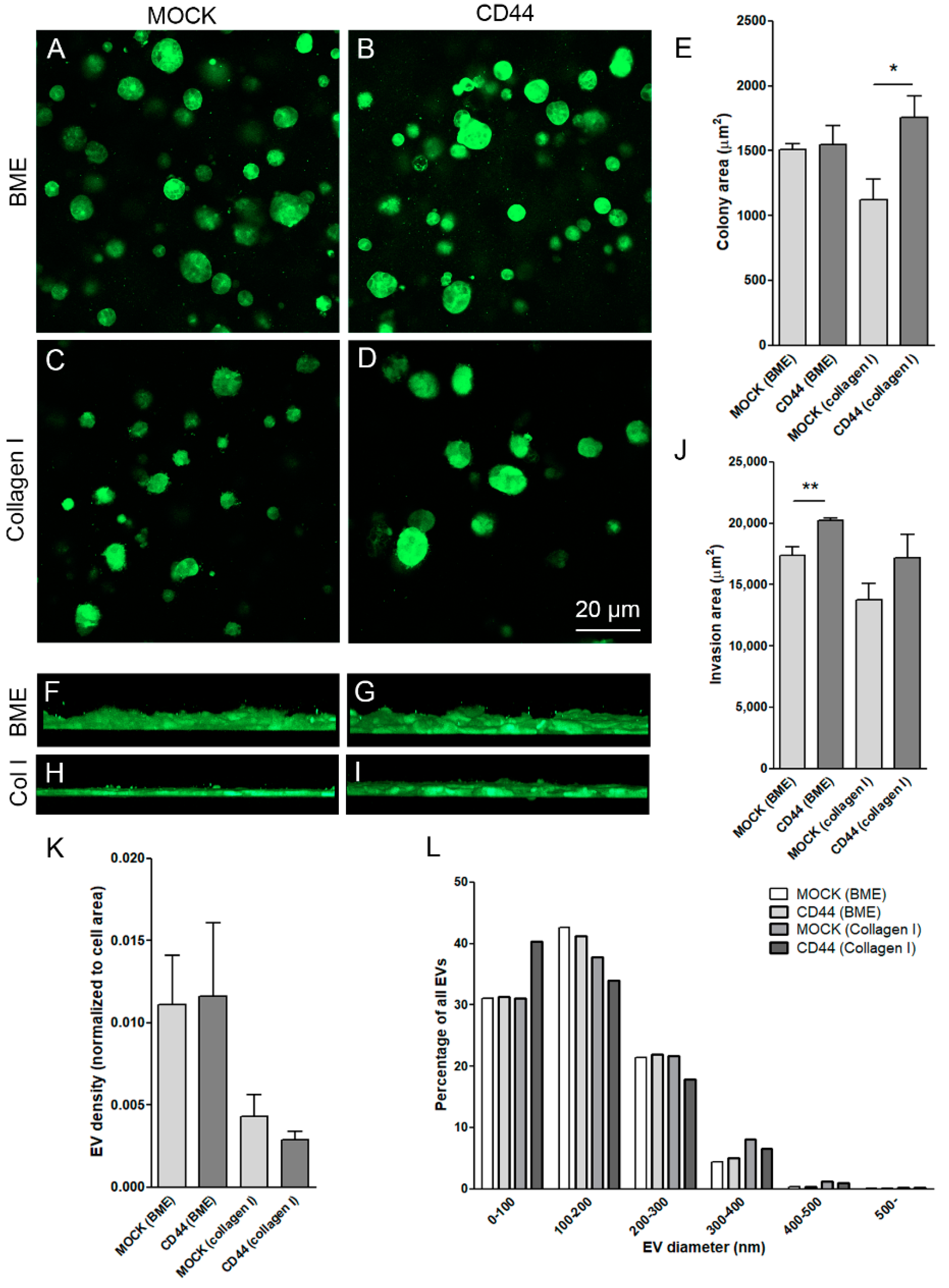
3.5. CD44s Induces Growth and Invasion but Not EV Secretion by MKN74 Cells in 3D Collagen Gels
3.6. CD44s Expression Does Not Affect the Growth and Other Features of CAM Tumors
3.7. CD44s Increases HA Content of the CAM Tumors
4. Discussion
4.1. Characterization of MNK74 Cells with CD44 Expression
4.2. Effect of CD44 Expression on HA Metabolism
4.3. Structure of HA Coat Induced by CD44
4.4. CD44-Expressing MKN74 Cells Secrete EVs That Carry CD44 and HA
4.5. Association of CD44 with Tumorigenic Properties
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BME | Basement membrane extract |
| CAM | Chick chorioallantoic membrane assay |
| CD44 | Cluster of differentiation 44 |
| ECM | Extracellular matrix |
| EMT | Epithelial-to-mesenchymal transition |
| EV | Extracellular vesicle |
| HA | Hyaluronan |
| bHABC | Biotinylated hyaluronan binding complex |
| HAS | Hyaluronan synthase |
| HYAL | Hyaluronidase |
| NTA | Nanoparticle tracking analysis |
References
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell 1989, 56, 1063–1072. [Google Scholar] [CrossRef]
- Aruffo, A.; Stamenkovic, I.; Melnick, M.; Underhill, C.B.; Seed, B. CD44 is the principal cell surface receptor for hyaluronate. Cell 1990, 61, 1303–1313. [Google Scholar] [CrossRef]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998, 140, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.W.; Isacke, C.M. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr. Biol. 1998, 8, 705–708. [Google Scholar] [CrossRef]
- Ghatak, S.; Misra, S.; Toole, B.P. Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. J. Biol. Chem. 2005, 280, 8875–8883. [Google Scholar] [CrossRef]
- Cho, S.H.; Park, Y.S.; Kim, H.J.; Kim, C.H.; Lim, S.W.; Huh, J.W.; Lee, J.H.; Kim, H.R. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 2012, 41, 211–218. [Google Scholar]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Sironen, R.K.; Tammi, M.; Tammi, R.; Auvinen, P.K.; Anttila, M.; Kosma, V.-M. Hyaluronan in human malignancies. Exp. Cell Res. 2011, 317, 383–391. [Google Scholar] [CrossRef]
- Naor, D.; Nedvetzki, S.; Golan, I.; Melnik, L.; Faitelson, Y. CD44 in Cancer. Crit. Rev. Clin. Lab. Sci. 2002, 39, 527–579. [Google Scholar] [CrossRef]
- Törrönen, K.; Nikunen, K.; Kärnä, R.; Tammi, M.; Tammi, R.; Rilla, K. Tissue distribution and subcellular localization of hyaluronan synthase isoenzymes. Histochem. Cell Biol. 2014, 141, 17–31. [Google Scholar] [CrossRef]
- Rilla, K.; Tiihonen, R.; Kultti, A.; Tammi, M.; Tammi, R. Pericellular hyaluronan coat visualized in live cells with a fluorescent probe is scaffolded by plasma membrane protrusions. J. Histochem. Cytochem. 2008, 56, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Evanko, S.P.; Potter-Perigo, S.; Johnson, P.Y.; Wight, T.N. Organization of hyaluronan and versican in the extracellular matrix of human fibroblasts treated with the viral mimetic poly I:C. J. Histochem. Cytochem. 2009, 57, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Jokela, T.A.; Kuokkanen, J.; Kärnä, R.; Pasonen-Seppänen, S.; Rilla, K.; Kössi, J.; Laato, M.; Tammi, R.H.; Tammi, M.I. Mannose reduces hyaluronan and leukocytes in wound granulation tissue and inhibits migration and hyaluronan-dependent monocyte binding. Wound Repair Regen. 2013, 21, 247–255. [Google Scholar] [CrossRef]
- Lin, W.; Shuster, S.; Maibach, H.I.; Stern, R. Patterns of hyaluronan staining are modified by fixation techniques. J. Histochem. Cytochem. 1997, 45, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Wickens, J.M.; Alsaab, H.O.; Kesharwani, P.; Bhise, K.; Amin, M.C.I.M.; Tekade, R.K.; Gupta, U.; Iyer, A.K. Recent advances in hyaluronic acid-decorated nanocarriers for targeted cancer therapy. Drug Discov. Today 2017, 22, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, D.; Padula, M.; Lu, J.; Jaiswal, R.; Djordjevic, S.; Bebawy, M. The role of CD44 and ERM proteins in expression and functionality of P-glycoprotein in breast cancer cells. Molecules 2016, 21, 290. [Google Scholar] [CrossRef] [PubMed]
- Arasu, U.T.; Kärnä, R.; Härkönen, K.; Oikari, S.; Koistinen, A.; Kröger, H.; Qu, C.; Lammi, M.J.; Rilla, K. Human mesenchymal stem cells secrete hyaluronan-coated extracellular vesicles. Matrix Biol. 2017, 64, 54–68. [Google Scholar] [CrossRef]
- Ramos, T.L.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef]
- Koistinen, V.; Härkönen, K.; Kärnä, R.; Arasu, U.T.; Oikari, S.; Rilla, K. EMT induced by EGF and wounding activates hyaluronan synthesis machinery and EV shedding in rat primary mesothelial cells. Matrix Biol. 2017, 63. [Google Scholar] [CrossRef] [PubMed]
- Rilla, K.; Pasonen-Seppänen, S.; Deen, A.J.; Koistinen, V.V.T.; Wojciechowski, S.; Oikari, S.; Kärnä, R.; Bart, G.; Törrönen, K.; Tammi, R.H.; et al. Hyaluronan production enhances shedding of plasma membrane-derived microvesicles. Exp. Cell Res. 2013, 319, 2006–2018. [Google Scholar] [CrossRef]
- Rilla, K.; Siiskonen, H.; Tammi, M.; Tammi, R. Hyaluronan-coated extracellular vesicles- A novel link between hyaluronan and cancer. Adv. Cancer Res. 2004, 123, 121–148. [Google Scholar]
- Hiltunen, E.L.J.; Anttila, M.; Kultti, A.; Ropponen, K.; Penttinen, J.; Yliskoski, M.; Kuronen, A.T.; Juhola, M.; Tammi, R.; Tammi, M.; et al. Elevated hyaluronan concentration without hyaluronidase activation in malignant epithelial ovarian tumors. Cancer Res. 2002, 62, 6410–6413. [Google Scholar]
- Tammi, R.; Rilla, K.; Pienimäki, J.-P.; MacCallum, D.K.; Hogg, M.; Luukkonen, M.; Hascall, V.C.; Tammi, M. Hyaluronan enters keratinocytes by a novel endocytic route for catabolism. J. Biol. Chem. 2001, 276, 35111–35122. [Google Scholar] [CrossRef]
- Hua, Q.; Knudson, C.B.; Knudson, W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J. Cell Sci. 1993, 106 Pt 1, 365–375. [Google Scholar]
- Rilla, K.; Siiskonen, H.; Spicer, A.P.; Hyttinen, J.M.T.; Tammi, M.I.; Tammi, R.H. Plasma membrane residence of hyaluronan synthase is coupled to its enzymatic activity. J. Biol. Chem. 2005, 280, 31890–31897. [Google Scholar] [CrossRef] [PubMed]
- Karalis, T.T.; Heldin, P.; Vynios, D.H.; Neill, T.; Buraschi, S.; Iozzo, R.V.; Karamanos, N.K.; Skandalis, S.S. Tumor-suppressive functions of 4-MU on breast cancer cells of different ER status: Regulation of hyaluronan/HAS2/CD44 and specific matrix effectors. Matrix Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Assmann, V.; Fieber, C.; Herrlich, P.; Hofmann, M.; Termeer, C.C.; Ahrens, T.; Simon, J.C. CD44 is the Principal mediator of hyaluronic-acid-induced melanoma cell proliferation. J. Investig. Dermatol. 2001, 116, 93–101. [Google Scholar] [CrossRef]
- Heldin, P.; Lin, C.-Y.; Kolliopoulos, C.; Chen, Y.-H.; Skandalis, S.S. Regulation of hyaluronan biosynthesis and clinical impact of excessive hyaluronan production. Matrix Biol. 2018. [Google Scholar] [CrossRef]
- Bano, F.; Banerji, S.; Howarth, M.; Jackson, D.G.; Richter, R.P. A single molecule assay to probe monovalent and multivalent bonds between hyaluronan and its key leukocyte receptor CD44 under force. Sci. Rep. 2016, 6, 34176. [Google Scholar] [CrossRef]
- Heldin, P.; Pertoft, H. Synthesis and assembly of the hyaluronan-containing coats around normal human mesothelial cells. Exp. Cell Res. 1993, 208, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Siiskonen, H.; Kärnä, R.; Hyttinen, J.M.; Tammi, R.H.; Tammi, M.I.; Rilla, K. Hyaluronan synthase 1 (HAS1) produces a cytokine-and glucose-inducible, CD44-dependent cell surface coat. Exp. Cell Res. 2014, 320, 153–163. [Google Scholar] [CrossRef]
- Siiskonen, H.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K. Hyaluronan synthase 1: A mysterious enzyme with unexpected functions. Front. Immunol. 2015, 6, 43. [Google Scholar] [CrossRef]
- Knudson, W.; Ishizuka, S.; Terabe, K.; Askew, E.B.; Knudson, C.B. The pericellular hyaluronan of articular chondrocytes. Matrix Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Twarock, S.; Tammi, M.I.; Savani, R.C.; Fischer, J.W. Hyaluronan stabilizes focal adhesions, filopodia, and the proliferative phenotype in esophageal squamous carcinoma cells. J. Biol. Chem. 2010, 285, 23276–23284. [Google Scholar] [CrossRef] [PubMed]
- Kultti, A.; Rilla, K.; Tiihonen, R.; Spicer, A.P.; Tammi, R.H.; Tammi, M.I. Hyaluronan synthesis induces microvillus-like cell surface protrusions. J. Biol. Chem. 2006, 281, 15821–15828. [Google Scholar] [CrossRef] [PubMed]
- Melero-Fernandez de Mera, R.M.; Arasu, U.T.; Kärnä, R.; Oikari, S.; Rilla, K.; Vigetti, D.; Passi, A.; Heldin, P.; Tammi, M.I.; Deen, A.J. Effects of mutations in the post-translational modification sites on the trafficking of hyaluronan synthase 2 (HAS2). Matrix Biol. 2018. [Google Scholar] [CrossRef]
- Nakamura, K.; Sawada, K.; Kinose, Y.; Yoshimura, A.; Toda, A.; Nakatsuka, E.; Hashimoto, K.; Mabuchi, S.; Morishige, K.; Kurachi, H.; et al. Exosomes promote ovarian cancer cell invasion through transfer of CD44 to peritoneal mesothelial cells. Mol. Cancer Res. 2017, 15, 78–92. [Google Scholar] [CrossRef]
- McDonald, B.; Kubes, P. Interactions between CD44 and hyaluronan in leukocyte trafficking. Front. Immunol. 2015, 6, 68. [Google Scholar] [CrossRef]
- Avigdor, A.; Goichberg, P.; Shivtiel, S.; Dar, A.; Peled, A.; Samira, S.; Kollet, O.; Hershkoviz, R.; Alon, R.; Hardan, I.; et al. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood 2004, 103, 2981–2989. [Google Scholar] [CrossRef]
- Richter, U.; Wicklein, D.; Geleff, S.; Schumacher, U. The interaction between CD44 on tumour cells and hyaluronan under physiologic flow conditions: Implications for metastasis formation. Histochem. Cell Biol. 2012, 137, 687–695. [Google Scholar] [CrossRef]
- Nandi, A.; Estess, P.; Siegelman, M.H. Hyaluronan anchoring and regulation on the surface of vascular endothelial cells is mediated through the functionally active form of CD44. J. Biol. Chem. 2000, 275, 14939–14948. [Google Scholar] [CrossRef]
- Koistinen, V.; Jokela, T.; Oikari, S.; Kärnä, R.; Tammi, M.; Rilla, K. Hyaluronan-positive plasma membrane protrusions exist on mesothelial cells in vivo. Histochem. Cell Biol. 2016, 145, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Qhattal, H.S.S.; Liu, X. Characterization of CD44-mediated cancer cell uptake and intracellular distribution of hyaluronan-grafted liposomes. Mol. Pharm. 2011, 8, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Jaracz, S.; Chen, J.; Kuznetsova, L.V.; Ojima, I. Recent advances in tumor-targeting anticancer drug conjugates. Bioorg. Med. Chem. 2005, 13, 5043–5054. [Google Scholar] [CrossRef] [PubMed]
- Oommen, O.P.; Duehrkop, C.; Nilsson, B.; Hilborn, J.; Varghese, O.P. Multifunctional hyaluronic acid and chondroitin sulfate nanoparticles: Impact of glycosaminoglycan presentation on receptor mediated cellular uptake and immune activation. ACS Appl. Mater. Interfaces 2016, 8, 20614–20624. [Google Scholar] [CrossRef]
- Wakamatsu, Y.; Sakamoto, N.; Oo, H.Z.; Naito, Y.; Uraoka, N.; Anami, K.; Sentani, K.; Oue, N.; Yasui, W. Expression of cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and lymph node metastasis of gastric cancer. Pathol. Int. 2012, 62, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Cao, D.; Jin, M.; Jia, Z.; Kong, F.; Ma, H.; Wang, Y.; Jiang, J. CD44 but not CD24 expression is related to poor prognosis in non-cardia adenocarcinoma of the stomach. BMC Gastroenterol. 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-W.; Chen, P.-C.; Zheng, C.-H.; Li, P.; Wang, J.-B.; Lin, J.-X.; Lu, J.; Chen, Q.-Y.; Cao, L.-L.; Lin, M.; et al. Evaluation of the prognostic value and functional roles of CD44v6 in gastric cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1809–1817. [Google Scholar] [CrossRef]
- Kodama, H.; Murata, S.; Ishida, M.; Yamamoto, H.; Yamaguchi, T.; Kaida, S.; Miyake, T.; Takebayashi, K.; Kushima, R.; Tani, M. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br. J. Cancer 2017, 116, 186–194. [Google Scholar] [CrossRef]
- Tan, P.; Yeoh, K.-G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology 2015, 149, 1153–1162. [Google Scholar] [CrossRef]
- Tammi, R.H.; Kultti, A.; Kosma, V.-M.; Pirinen, R.; Auvinen, P.; Tammi, M.I. Hyaluronan in human tumors: Pathobiological and prognostic messages from cell-associated and stromal hyaluronan. Semin. Cancer Biol. 2008, 18, 288–295. [Google Scholar] [CrossRef]
- Auvinen, P.; Tammi, R.; Kosma, V.-M.; Sironen, R.; Soini, Y.; Mannermaa, A.; Tumelius, R.; Uljas, E.; Tammi, M. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int. J. Cancer 2013, 132, 531–539. [Google Scholar] [CrossRef]
- Jeong, B.-J.; Liang, Z.L.; Huang, S.M.; Lim, J.S.; Kim, J.M.; Lee, H.J. CD44 is associated with tumor recurrence and is an independent poor prognostic factor for patients with localized clear cell renal cell carcinoma after nephrectomy. Exp. Ther. Med. 2012, 3, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, S.; Anttila, M.A.; Voutilainen, K.; Tammi, R.H.; Tammi, M.I.; Saarikoski, S.V.; Kosma, V.-M. CD44 expression indicates favorable prognosis in epithelial ovarian cancer. Clin. Cancer Res. 2003, 9, 5318–5324. [Google Scholar]
- Matuura, H.; Miyamoto, M.; Takano, M.; Soyama, H.; Aoyama, T.; Yoshikawa, T.; Kato, K.; Sakamoto, T.; Kuwahara, M.; Takasaki, K.; et al. Low expression of CD44 is an independent factor of poor prognosis in ovarian mucinous carcinoma. Anticancer Res. 2018, 38, 717–722. [Google Scholar]
- Schmits, R.; Filmus, J.; Gerwin, N.; Senaldi, G.; Kiefer, F.; Kundig, T.; Wakeham, A.; Shahinian, A.; Catzavelos, C.; Rak, J.; et al. CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity. Blood 1997, 90, 2217–2233. [Google Scholar] [PubMed]
- Iozzo, R.V.; Gubbiotti, M.A. Extracellular matrix: The driving force of mammalian diseases. Matrix Biol. 2018, 71–72, 1–9. [Google Scholar] [CrossRef]
- Xia, P.; Xu, X.-Y. Prognostic significance of CD44 in human colon cancer and gastric cancer: Evidence from bioinformatic analyses. Oncotarget 2016, 7, 45538–45546. [Google Scholar] [CrossRef] [PubMed]
- Tammi, M.I.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated hyaluronan metabolism in the tumor matrix—Causes and consequences. Matrix Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rilla, K.; Mustonen, A.-M.; Arasu, U.T.; Härkönen, K.; Matilainen, J.; Nieminen, P. Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol. 2017, 75–76, 201–219. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequences | ||
|---|---|---|---|
| Hyaluronan synthase 1 | HAS1 | Forward 5′ | CAAGATTCTTCAGTCTGGAC |
| Reverse 5′ | TAAGAACGAGGAGAAAGCAG | ||
| Hyaluronan synthase 2 | HAS2 | Forward 5′ | CAGAATCCAAACAGACAGTTC |
| Reverse 5′ | TAAGGTGTTGTGTGTGACTG | ||
| Hyaluronan synthase 3 | HAS3 | Forward 5′ | CTTAAGGGTTGCTTGCTTGC |
| Reverse 5′ | GTTCGTGGGAGATGAAGGAA | ||
| CD44 | CD44 | Forward 5′ | CATCTACCCCAGCAACCCTA |
| Reverse 5′ | CTGTCTGTGCTGTCGGTGAT | ||
| Hyaluronidase 2 | HYAL2 | Forward 5′ | CCTCTGGGGCTTCTACCTCT |
| Reverse 5′ | CTGAACACGGAAGCTCACAA | ||
| Hyaluronidase 3 | HYAL3 | Forward 5′ | GCTGGCATAGTATGGCTTCC |
| Reverse 5′ | ACACCAATGGACTGCACAAG | ||
| Ribosomal protein, Large, P0 | RPLP0 | Forward 5′ | AGATGCAGCAGATCCGCAT |
| Reverse 5′ | GTGGTGATACCTAAAGCCTG | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Härkönen, K.; Oikari, S.; Kyykallio, H.; Capra, J.; Hakkola, S.; Ketola, K.; Thanigai Arasu, U.; Daaboul, G.; Malloy, A.; Oliveira, C.; et al. CD44s Assembles Hyaluronan Coat on Filopodia and Extracellular Vesicles and Induces Tumorigenicity of MKN74 Gastric Carcinoma Cells. Cells 2019, 8, 276. https://doi.org/10.3390/cells8030276
Härkönen K, Oikari S, Kyykallio H, Capra J, Hakkola S, Ketola K, Thanigai Arasu U, Daaboul G, Malloy A, Oliveira C, et al. CD44s Assembles Hyaluronan Coat on Filopodia and Extracellular Vesicles and Induces Tumorigenicity of MKN74 Gastric Carcinoma Cells. Cells. 2019; 8(3):276. https://doi.org/10.3390/cells8030276
Chicago/Turabian StyleHärkönen, Kai, Sanna Oikari, Heikki Kyykallio, Janne Capra, Sini Hakkola, Kirsi Ketola, Uma Thanigai Arasu, George Daaboul, Andrew Malloy, Carla Oliveira, and et al. 2019. "CD44s Assembles Hyaluronan Coat on Filopodia and Extracellular Vesicles and Induces Tumorigenicity of MKN74 Gastric Carcinoma Cells" Cells 8, no. 3: 276. https://doi.org/10.3390/cells8030276





