1. Introduction
Understanding the genetic basis and the driving forces of mate choice in animals has always been a major goal of evolutionary ecologists [
1,
2]. It has been proposed that females prefer males who can maximize their reproductive success and increase offspring quality/fitness [
3,
4]. Immunocompetence is undoubtedly an essential index of an individual’s fitness [
5,
6] and major histocompatibility complex (MHC) genes are suitable candidates in investigating the genetic basis underlying mate choice decisions, as MHC molecules can recognize and present antigens to T cells and trigger immune reactions [
7,
8,
9,
10]. A growing body of evidence shows the association between pathogen resistance and MHC haplotypes or alleles [
11,
12,
13,
14].
In recent years, an increasing number of studies have focused on MHC-associated mate choice, including three non-exclusive hypotheses that could explain the MHC-based mate choice. Firstly, heterozygous advantage: according to this hypothesis, the choosy sex could obtain additive benefits from mating with MHC-heterozygous mates whose disease resistance might be inherited by their offspring [
15,
16]. For example, in tuco-tucos (
Ctenomys spp.), females prefer MHC-heterozygous males [
17]. In the scarlet rosefinch (
Carpodacus erythrinus), social males with low MHC heterozygosity are cheated on by their females more frequently than highly MHC-heterozygous males [
18]. Secondly, the genetic compatibility hypothesis: the choosy sex is assumed to select MHC-dissimilar partners, resulting in the production of offspring with diverse genotypes that can recognize a broad array of pathogens and hence increase their fitness [
15,
16,
19]. In the grey mouse lemur (
Microcebus murinus), the fathers have lower allele sharing and a greater amino acid distance to the mother than the randomly assigned males [
20]. A study on blue petrels (
Halobaena caerulea) revealed that females mated more with functionally (not evolutionary) MHC-dissimilar males than with random males [
21]. Similar patterns have been observed in great frigatebirds (
Fregata minor) [
22], the Chinese rose bitterling (
Rhodeus ocellatus) [
23], the pot-bellied seahorse (
Hippocampus abdominalis) [
24], the fat-tailed dwarf lemur (
Cheirogaleus medius) [
25], and mandrills (
Mandrillus spp.) [
26]. Furthermore, to acquire the best immunogenetic composition for their offspring, females would choose mates with the most appropriate MHC diversity (maximum or intermediate), which is also referred to as the optimal hypothesis, and it is an extension of the genetic compatibility hypothesis [
10]. Studies in sticklebacks (family Gasterosteidae) have found that females with many alleles prefer males with few alleles, and vice versa, in order to obtain an optimal level of MHC diversity in their offspring for resistance against parasites and pathogens [
27,
28]. Thirdly, the inbreeding avoidance hypothesis: the choosy sex is expected to seek dissimilar partners, not only with regard to MHC, but also genome-wide, in order to gain fitness benefits [
29,
30].
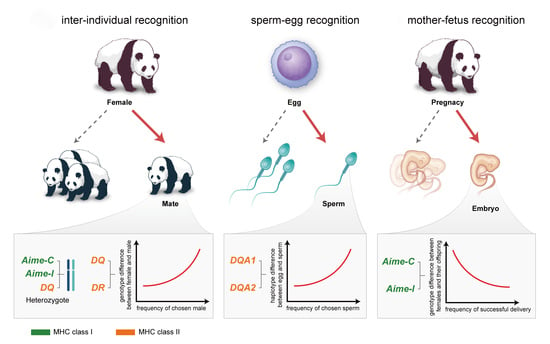
Mate choice results from non-random reproductive investment, which could happen at the precopulatory stage or/and at the postcopulatory stage. In the precopulatory stage (or individual recognition level), the choosy sex uses visual, acoustic, or odor cues to choose mates [
15]. In the postcopulatory stage, the females increase offspring quality/fitness by their eggs differentiating between sperm during fertilization (sperm-egg recognition), and by following a differential allocation strategy during embryo implantation (mother-fetus recognition) [
16]. Human leukocyte antigen (HLA) class I and class II molecules are expressed on sperm cell surfaces [
31,
32,
33], making the complementary cryptic female choice for MHC genotypes possible at the gamete level. Sperm selection that targets different levels of MHC diversity has been reported in several species. In some rodent species, females refuse to accept MHC-similar sperm [
34], and in humans, females that mate with a male who has the same HLA haplotype tend to have a greater chance of spontaneous abortion [
35,
36,
37]. In red junglefowl (
Gallus gallus), the eggs favor sperm that is from MHC-dissimilar males [
38]. In contrast, a fertilization advantage for MHC-similar mates has been observed in Atlantic salmon (
Salmo salar) [
39], Chinook salmon (
Oncorhynchus tshawytscha) [
40], and guppies (
Poecilia reticulata) [
41]. A recent study of the three-spined stickleback (
Gasterosteus aculeatus) revealed that sperm selection resulted in offspring with an intermediate level of MHC diversity [
42].
The giant panda (
Ailuropoda melanoleuca) is an endangered species and its captive breeding has always been focused in China. Giant pandas usually have polyandrous/polygynous multiple mating systems [
43]. Females are very choosy, and they have significantly higher copulation and birth rates when paired with preferred males [
44]. In breeding programs, natural mating as well as artificial insemination are adopted to increase reproductive success [
45]. Although the number of female giant pandas that are available to breed has increased, the fertilization rate is still low (~50% for nearly 20 years). Furthermore, female giant pandas have spontaneous abortions when they are inseminated by sperm from a male that they do not like [
46]. Therefore, there may be an MHC-based mechanism that determines mate choice and fertilization in giant pandas.
It is best to investigate all MHC genes concerning mate choice, as MHC class I and class II molecules mainly present intracellular and extracellular pathogen-derived antigens, respectively [
9], and the selection of MHC genes might influence mate choice results [
47]. Few studies have focused on a large region of the MHC due to a lack of structural knowledge and an effective genotyping method [
48], including a giant panda mate choice study that only used three MHC class II genes [
49]. In previous studies, we characterized six functional MHC class II genes (
Aime-DRA,
Aime-DRB3,
Aime-DQA1,
Aime-DQA2,
Aime-DQB1, and
Aime-DQB2) [
50,
51] and four classical MHC class I genes (
Aime-C,
Aime-F,
Aime-I, and
Aime-L) [
52], and we developed their genotyping protocols in the giant panda. Therefore, our previous studies provide a good foundation to investigate the relationship between a large number of MHC genes and female choice at the inter-individual, sperm-egg, and mother-fetus recognition levels in giant pandas.
In the present study, we took advantage of multiple years of observations and the possession of genetic data of a captive population in the Wolong Chinese Research and Conservation Center for the Giant Panda. We aimed to: (1) test three mate choice hypotheses at the individual recognition level (MHC-heterozygous choice, MHC-compatibility choice, and inbreeding avoidance) while using 10 MHC genes and seven microsatellites. If the heterozygote advantage is the main driving force of female mate choice, more diverse males should be preferred than less diverse ones, regardless of the females’ MHC genotypes. If female choice favors the production of offspring with high fitness that is based on MHC compatibility, we expect that females would choose MHC-dissimilar partners that provide the most appropriate level of MHC dissimilarity (maximum or intermediate) in the offspring. If mate choice aims to avoid inbreeding, then we expect genome-wide dissimilarity between partners (MHC and microsatellites); (2) test whether cryptic female choice for MHC compatibility occurs at the gamete level; and, (3) characterize successful embryo implantations by comparing the MHC genotypes of mothers and their offspring.
4. Discussion
4.1. Female Choice at the Inter-Individual Recognition Level
We found that female giant pandas preferred males with MHC-heterozygous and MHC-dissimilar genotypes, which supports the heterozygosity advantage and disassortative choice of the compatibility hypotheses, respectively. It has been proposed that females usually use male ornaments as a cue for choosing MHC-heterozygous males, and olfaction as a cue for choosing MHC-dissimilar males [
15]. Body color varies among giant pandas, and it may reflect condition [
73,
74,
75]. Therefore, we propose that female giant pandas assess males’ physical condition in order to choose MHC-heterozygous mates by evaluating the depth of body color. During the mating season, giant pandas secrete musk from their anal glands and in their urine, which is highly odorous [
43], thus it is possible that females use odor to choose among male MHC genotypes. Animals can simultaneously use two cues, and the two strategies are not mutually exclusive, particularly in species with both good vision and olfaction [
19,
76,
77]. A recent study on giant pandas reported multimodal signal behavior between females and males, including olfaction, vision, and hearing [
55], supporting the use of two strategies. It has been reported that female pandas use odor cues prior to face-to-face meetings, so we propose that females choose MHC-dissimilar mating partners based on odor cues alone, and they use body color depth to visually choose MHC-heterozygous males. A “multiple-cue strategy” was also found in a study of female mice that changed strategy when the males’ urinary scent-marking rate changed [
78], and in lizards that use coloration as a mate choice cue at long distances, but use odor at short distances [
79]. Using multiple cues to choose a mate may be more common than expected, as females gain more information and reduce their selection costs [
80].
The preference for MHC-dissimilar mates might be due to inbreeding avoidance in giant panda populations. In general, if mate choice aims to avoid inbreeding, then we expected to see significantly lower relatedness in the observed pairs than in randomly assigned pairs [
20,
63,
81]. However, our results revealed no significant difference between observed pairs and randomly assigned pairs (
Table 1), suggesting that female giant pandas do not attempt to avoid inbreeding. Avoiding inbreeding may not be as important to females as maximizing the number of MHC polymorphisms in their offspring in order to resist pathogens. In addition, we did not find any evidence for the heterozygote advantage hypothesis that is based on the MLH and
d2 results, suggesting that overall genetic diversity has no effect on mate choice. These findings indicate that female mate choice targets functional MHC genes rather than other regions, or it is a byproduct of inbreeding avoidance.
Some studies have reported that giant pandas are susceptible to parasites and viruses [
82,
83,
84,
85], suggesting that the MHC genes are important in female mate choice at the inter-individual recognition level. Furthermore, our results show that females favored partners that were the most MHC-dissimilar to themselves, resulting in offspring with high heterozygosity. Whether offspring have high immunocompetence should be addressed in future studies regarding the relationship between MHC heterozygosity and immunocompetence in the giant panda.
4.2. Cryptic Female Choice at the Sperm-Egg Recognition Level
Female giant pandas usually mate with multiple males, possibly to increase fertilization success and offspring genetic quality, as has been found in other species [
3,
86,
87]. Our results revealed that sperm and eggs do not randomly combine, and that sperm from zygotes observed was more dissimilar to eggs at DQA1 and DQA2 than sperm from other zygotes (
Figure 7 and
Figure 8 and
Table 5). Two mechanisms may explain this: cryptic female choice or sperm competition [
15,
16,
20], but we could not identify which was the most important. Nevertheless, combined sperm and eggs had the maximum functional amino acid distance, which supports the female MHC-disassortative choice of compatibility hypothesis that is described above.
4.3. Mother-Fetus Recognition Level
The observed zygotes were more similar to mothers at
Aime-C and
Aime-I than the randomly assigned zygotes and other zygotes (
Figure 9). These findings suggest that the observed zygotes had higher compatibility at
Aime-C and
Aime-I, which could decrease or block graft rejections from the mother’s immune system. MHC class I molecules play an important role in the immune reaction between the mother and fetus, e.g., HLA-G molecules are present at the mother-child interface of trophoblastic cells and protect the fetus from the lytic activity of maternal uterine natural killer cells [
88,
89]. The identification of an HLA-G ortholog in the giant panda would elucidate the effects of mother-fetus immunity in this species; however, MHC class I genes have similar loci, making HLA-G orthologs difficult to identify in the giant panda.
4.4. Hierarchical and Cooperative Effects
From the super haplotype level to individual loci, female mate choice was hierarchical in nature. For example, we found that SuHa, which represents all of the functional MHC genes in giant pandas, predicted MHC-heterozygous female choice. However, when separately analyzing SuHaI and SuHaII, the effect could only be observed in SuHaII. When we then excluded DR from SuHaII and analyzed DQ alone, the heterozygosity advantage could still be detected. In contrast, the heterozygosity advantage could not be detected when only analyzing DR. Finally, when we focused on the individual locus level, only one out of three DQ genes (DQB1) still predicted female choice. A similar pattern was found for the genetic compatibility hypothesis.
Alternatively, multiple MHC genes that act cooperatively can explain the above example. The effect of DQ seemed to be larger than that of DQB1 alongside DQA1 and DQA2. The integration of the loci or super haplotypes had a greater effect than the sum of the loci or super haplotypes.
The SuHa results were inconsistent with those of SuHaII with respect to inter-individual recognition, suggesting that SuHaII is more important than SuHaI. This may have been caused by greater variation in SuHaII than in SuHaI.
4.5. Relative Importance of MHC-I, MHC-II, DQ, and DR
MHC class I, MHC class II, DQ, and DR were differentially important at the inter-individual, gamete, and mother-fetus recognition levels. More MHC II genes predicted compatible female mate choice than the MHC I genes, e.g., naturally mated males were more dissimilar to their partners at SuHaII but not at SuHaI. Furthermore, all four of the MHC class II genes (DQA1, DQA2, DQB1, and DRB3) predicted compatible female mate choice, while only one out of three MHC class I genes had a significant result. At the gamete level, the sperm were more dissimilar to eggs at DQ than at DR. At the mother-fetus recognition level, the observed zygotes (offspring) were more similar to their mothers at MHC class I genes, but not at MHC class II genes.
Genes that predict female mate choice or gamete selection may be more important than other MHC loci that are involved in pathogen resistance, as revealed by many studies in which distinct molecules that are coded by different alleles recognize specific pathogens, and their ability to resist pathogens differs [
11,
12,
13,
14], such as the DQ region. The DQ region in the giant panda differs to that in other mammals, because it contains more genes and alleles than the DR region [
59]. DRB3 exhibited the most variation among the seven individual polymorphic loci. MHC polymorphisms may be driven by sexual selection [
10], which is in line with our results at DQ and DRB3.
Our findings suggest that MHC loci do not play equal roles in female mate choice at the inter-individual recognition level or at other levels, and that targeted MHC loci may be key for female mate choice at the individual recognition level. Therefore, more MHC loci should be isolated and large MHC regions surveyed.