Involvement of CXCR4 in Normal and Abnormal Development
Abstract
:1. Introduction
2. CXCR4 Involvement in Normal Development
2.1. CXCR4 Knock-Out Studies
2.2. SDF-1/CXCR4 Functions in Tissue Repair
3. CXCR4 Involvement in Abnormal Pathological Development
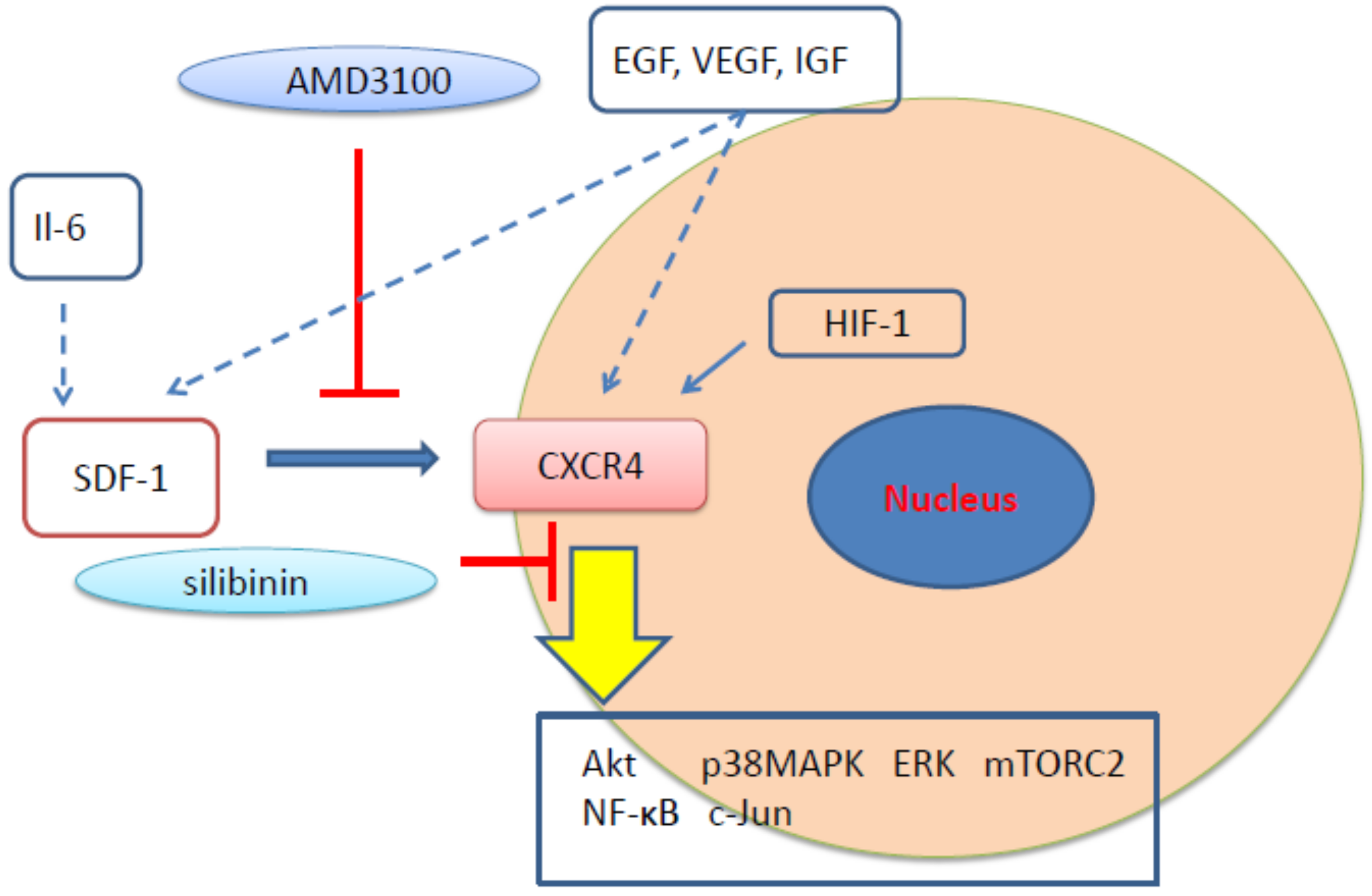
3.1. CXCR4 in Cancer
3.2. Pulmonary Arterial Hypertension (PAH)
3.3. WHIM Syndrome
4. CXCR4 Involvement in Normal and Abnormal Development
CXCR4 Signal Transduction and the CXCR4 Family
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Balestrieri, M.L.; Balestrieri, A.; Mancini, F.P.; Napoli, C. Understanding the immunoangiostatic cxc chemokine network. Cardiovasc. Res. 2008, 78, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Pawig, L.; Klasen, C.; Weber, C.; Bernhagen, J.; Noels, H. Diversity and inter-connections in the cxcr4 chemokine receptor/ligand family: Molecular perspectives. Front. Immunol. 2015, 6, 429. [Google Scholar] [CrossRef] [PubMed]
- Imitola, J.; Raddassi, K.; Park, K.I.; Mueller, F.J.; Nieto, M.; Teng, Y.D.; Frenkel, D.; Li, J.; Sidman, R.L.; Walsh, C.A.; et al. Directed migration of neural stem cells to sites of cns injury by the stromal cell-derived factor 1alpha/cxc chemokine receptor 4 pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 18117–18122. [Google Scholar] [CrossRef] [PubMed]
- Doring, Y.; Pawig, L.; Weber, C.; Noels, H. The cxcl12/cxcr4 chemokine ligand/receptor axis in cardiovascular disease. Front. Physiol. 2014, 5, 212. [Google Scholar] [PubMed]
- Askari, A.T.; Unzek, S.; Popovic, Z.B.; Goldman, C.K.; Forudi, F.; Kiedrowski, M.; Rovner, A.; Ellis, S.G.; Thomas, J.D.; DiCorleto, P.E.; et al. Effect of stromal-cell-derived factor 1 on stem-cell homing and tissue regeneration in ischaemic cardiomyopathy. Lancet 2003, 362, 697–703. [Google Scholar] [CrossRef]
- Sundararaman, S.; Miller, T.J.; Pastore, J.M.; Kiedrowski, M.; Aras, R.; Penn, M.S. Plasmid-based transient human stromal cell-derived factor-1 gene transfer improves cardiac function in chronic heart failure. Gene Ther. 2011, 18, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Penn, M.S.; Pastore, J.; Miller, T.; Aras, R. Sdf-1 in myocardial repair. Gene Ther. 2012, 19, 583–587. [Google Scholar] [CrossRef]
- Unzek, S.; Zhang, M.; Mal, N.; Mills, W.R.; Laurita, K.R.; Penn, M.S. Sdf-1 recruits cardiac stem cell-like cells that depolarize in vivo. Cell Transplant. 2007, 16, 879–886. [Google Scholar] [CrossRef]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef]
- Schunkert, H.; Konig, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef]
- Debnath, B.; Xu, S.; Grande, F.; Garofalo, A.; Neamati, N. Small molecule inhibitors of cxcr4. Theranostics 2013, 3, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Chen, S.; Wang, S.; Yu, X.; Chen, Y.; Jiang, M.; Zhuang, K.; Ho, W.; Hou, W.; Huang, J.; et al. Genome editing of cxcr4 by crispr/cas9 confers cells resistant to hiv-1 infection. Sci. Rep. 2015, 5, 15577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, S.; Xiao, Q.; Liu, Z.; Liu, S.; Hou, P.; Zhou, L.; Hou, W.; Ho, W.; Li, C.; et al. Genome modification of cxcr4 by staphylococcus aureus cas9 renders cells resistance to hiv-1 infection. Retrovirology 2017, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome editing of the hiv co-receptors ccr5 and cxcr4 by crispr-cas9 protects cd4(+) t cells from hiv-1 infection. Cell Biosci. 2017, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Oberlin, E.; Amara, A.; Bachelerie, F.; Bessia, C.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Schwartz, O.; Heard, J.M.; Clark-Lewis, I.; Legler, D.F.; et al. The cxc chemokine sdf-1 is the ligand for lestr/fusin and prevents infection by t-cell-line-adapted hiv-1. Nature 1996, 382, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Bleul, C.C.; Farzan, M.; Choe, H.; Parolin, C.; Clark-Lewis, I.; Sodroski, J.; Springer, T.A. The lymphocyte chemoattractant sdf-1 is a ligand for lestr/fusin and blocks hiv-1 entry. Nature 1996, 382, 829–833. [Google Scholar] [CrossRef]
- Nagasawa, T.; Nakajima, T.; Tachibana, K.; Iizasa, H.; Bleul, C.C.; Yoshie, O.; Matsushima, K.; Yoshida, N.; Springer, T.A.; Kishimoto, T. Molecular cloning and characterization of a murine pre-b-cell growth-stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc. Natl. Acad. Sci. USA 1996, 93, 14726–14729. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, T.; Tachibana, K.; Kishimoto, T. A novel cxc chemokine pbsf/sdf-1 and its receptor cxcr4: Their functions in development, hematopoiesis and hiv infection. Semin. Immunol. 1998, 10, 179–185. [Google Scholar] [CrossRef]
- Zou, Y.R.; Kottmann, A.H.; Kuroda, M.; Taniuchi, I.; Littman, D.R. Function of the chemokine receptor cxcr4 in haematopoiesis and in cerebellar development. Nature 1998, 393, 595–599. [Google Scholar] [CrossRef]
- Nagasawa, T.; Hirota, S.; Tachibana, K.; Takakura, N.; Nishikawa, S.; Kitamura, Y.; Yoshida, N.; Kikutani, H.; Kishimoto, T. Defects of b-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the cxc chemokine pbsf/sdf-1. Nature 1996, 382, 635–638. [Google Scholar] [CrossRef]
- Tachibana, K.; Hirota, S.; Iizasa, H.; Yoshida, H.; Kawabata, K.; Kataoka, Y.; Kitamura, Y.; Matsushima, K.; Yoshida, N.; Nishikawa, S.; et al. The chemokine receptor cxcr4 is essential for vascularization of the gastrointestinal tract. Nature 1998, 393, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Wunderlin, M.; Spieth, K.; Knochel, W.; Gierschik, P.; Moepps, B. Xenopus laevis stromal cell-derived factor 1: Conservation of structure and function during vertebrate development. J. Immunol. 2002, 168, 2340–2347. [Google Scholar] [CrossRef]
- Zhang, Z.; Cerrato, F.; Xu, H.; Vitelli, F.; Morishima, M.; Vincentz, J.; Furuta, Y.; Ma, L.; Martin, J.F.; Baldini, A.; et al. Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development 2005, 132, 5307–5315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huynh, T.; Baldini, A. Mesodermal expression of tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development 2006, 133, 3587–3595. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.S.; Werling, U.; Braunstein, E.M.; Liao, J.; Nowotschin, S.; Edelmann, W.; Hebert, J.M.; Morrow, B.E. Inactivation of tbx1 in the pharyngeal endoderm results in 22q11ds malformations. Development 2006, 133, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, H.; Srivastava, D. Unraveling the genetic and developmental mysteries of 22q11 deletion syndrome. Trends Mol. Med. 2003, 9, 383–389. [Google Scholar] [CrossRef]
- Olesnicky Killian, E.C.; Birkholz, D.A.; Artinger, K.B. A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev. Biol. 2009, 333, 161–172. [Google Scholar] [CrossRef]
- Kim, B.G.; Kim, Y.H.; Stanley, E.L.; Garrido-Martin, E.M.; Lee, Y.J.; Oh, S.P. Cxcl12-cxcr4 signalling plays an essential role in proper patterning of aortic arch and pulmonary arteries. Cardiovasc. Res. 2017, 113, 1677–1687. [Google Scholar] [CrossRef]
- Escot, S.; Blavet, C.; Faure, E.; Zaffran, S.; Duband, J.L.; Fournier-Thibault, C. Disruption of cxcr4 signaling in pharyngeal neural crest cells causes digeorge syndrome-like malformations. Development 2016, 143, 582–588. [Google Scholar] [CrossRef]
- Page, M.; Ridge, L.; Gold Diaz, D.; Tsogbayar, T.; Scambler, P.J.; Ivins, S. Loss of cxcl12/cxcr4 signalling impacts several aspects of cardiovascular development but does not exacerbate tbx1 haploinsufficiency. PLoS ONE 2018, 13, e0207251. [Google Scholar] [CrossRef]
- Zhang, S.J.; Song, X.Y.; He, M.; Yu, S.B. Effect of tgf-beta1/sdf-1/cxcr4 signal on bm-mscs homing in rat heart of ischemia/perfusion injury. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 899–905. [Google Scholar] [PubMed]
- Zisa, D.; Shabbir, A.; Mastri, M.; Taylor, T.; Aleksic, I.; McDaniel, M.; Suzuki, G.; Lee, T. Intramuscular vegf activates an sdf1-dependent progenitor cell cascade and an sdf1-independent muscle paracrine cascade for cardiac repair. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2422–H2432. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Ong, L.L.; Furlani, D.; Kaminski, A.; Liebold, A.; Lutzow, K.; Lendlein, A.; Wang, J.; Li, R.K.; et al. Localized sdf-1alpha gene release mediated by collagen substrate induces cd117 stem cells homing. J. Cell. Mol. Med. 2010, 14, 392–402. [Google Scholar] [CrossRef]
- Hatzistergos, K.E.; Saur, D.; Seidler, B.; Balkan, W.; Breton, M.; Valasaki, K.; Takeuchi, L.M.; Landin, A.M.; Khan, A.; Hare, J.M. Stimulatory effects of mesenchymal stem cells on ckit+ cardiac stem cells are mediated by sdf1/cxcr4 and scf/ckit signaling pathways. Circ. Res. 2016, 119, 921–930. [Google Scholar] [CrossRef]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef]
- Ellison, G.M.; Vicinanza, C.; Smith, A.J.; Aquila, I.; Leone, A.; Waring, C.D.; Henning, B.J.; Stirparo, G.G.; Papait, R.; Scarfo, M.; et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 2013, 154, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, E.; Hosoda, T. How do resident stem cells repair the damaged myocardium? World J. Stem Cells 2015, 7, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Zhang, L.; Yan, J.; Chen, J.; Cai, W.; Razzaque, S.; Jeong, D.; Sheng, W.; Bu, L.; Xu, M.; et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat. Commun. 2015, 6, 8701. [Google Scholar] [CrossRef] [PubMed]
- Keith, M.C.; Bolli, R. "String theory" of c-kit(pos) cardiac cells: A new paradigm regarding the nature of these cells that may reconcile apparently discrepant results. Circ. Res. 2015, 116, 1216–1230. [Google Scholar] [CrossRef]
- Kawaguchi, N.; Smith, A.J.; Waring, C.D.; Hasan, M.K.; Miyamoto, S.; Matsuoka, R.; Ellison, G.M. C-kitpos gata-4 high rat cardiac stem cells foster adult cardiomyocyte survival through igf-1 paracrine signalling. PLoS ONE 2010, 5, e14297. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kawaguchi, N.; Ellison, G.M.; Matsuoka, R.; Shin’oka, T.; Kurosawa, H. Characterization of long-term cultured c-kit+ cardiac stem cells derived from adult rat hearts. Stem Cells Dev. 2010, 19, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Alshammary, S.; Fukushima, S.; Miyagawa, S.; Matsuda, T.; Nishi, H.; Saito, A.; Kamata, S.; Asahara, T.; Sawa, Y. Impact of cardiac stem cell sheet transplantation on myocardial infarction. Surg. Today 2013, 43, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, N.; Nakanishi, T. Cardiomyocyte regeneration. Cells 2013, 2, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Zuo, K.; Kuang, D.; Wang, Y.; Xia, Y.; Tong, W.; Wang, X.; Chen, Y.; Duan, Y.; Wang, G. Scf/c-kit transactivates cxcr4-serine 339 phosphorylation through g protein-coupled receptor kinase 6 and regulates cardiac stem cell migration. Sci. Rep. 2016, 6, 26812. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.; Ling, T.Y.; Lin, H.Y.; Liou, J.T.; Liu, F.C.; Chen, I.C.; Lee, S.W.; Hsu, Y.; Lai, D.M. Sdf-1/cxcr4 signaling maintains stemness signature in mouse neural stem/progenitor cells. Stem Cells Int. 2017, 2017, 2493752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hua, Q.; Tang, K.; Shi, C.; Xie, X.; Zhang, R. Cxcr4 activation promotes differentiation of human embryonic stem cells to neural stem cells. Neuroscience 2016, 337, 88–97. [Google Scholar] [CrossRef]
- Rafii, S.; Cao, Z.; Lis, R.; Siempos, I.I.; Chavez, D.; Shido, K.; Rabbany, S.Y.; Ding, B.S. Platelet-derived sdf-1 primes the pulmonary capillary vascular niche to drive lung alveolar regeneration. Nat. Cell Biol. 2015, 17, 123–136. [Google Scholar] [CrossRef]
- Lefrancais, E.; Ortiz-Munoz, G.; Caudrillier, A.; Mallavia, B.; Liu, F.; Sayah, D.M.; Thornton, E.E.; Headley, M.B.; David, T.; Coughlin, S.R.; et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 2017, 544, 105–109. [Google Scholar] [CrossRef]
- Borges, I.; Sena, I.; Azevedo, P.; Andreotti, J.; Almeida, V.; Paiva, A.; Santos, G.; Guerra, D.; Prazeres, P.; Mesquita, L.L.; et al. Lung as a niche for hematopoietic progenitors. Stem Cell Rev. 2017, 13, 567–574. [Google Scholar] [CrossRef]
- Sawitza, I.; Kordes, C.; Reister, S.; Haussinger, D. The niche of stellate cells within rat liver. Hepatology 2009, 50, 1617–1624. [Google Scholar] [CrossRef]
- Furusato, B.; Mohamed, A.; Uhlen, M.; Rhim, J.S. Cxcr4 and cancer. Pathol. Int. 2010, 60, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, C.; Xu, W.; Zhang, J.; Zheng, Y.; Lu, Z.; Liu, D.; Jiang, K. Cxc motif chemokine receptor 4 gene polymorphism and cancer risk. Medicine 2016, 95, e5317. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.V.; Short, S.P.; Neel, N.F.; Salvo, V.A.; Zhu, Y.; Elliott, S.; Wei, Y.; Yu, D.; Sun, M.; Muir, S.E.; et al. Cytokine receptor cxcr4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011, 71, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Neel, N.F.; Schutyser, E.; Raman, D.; Richmond, A. Deletion of the cooh-terminal domain of cxc chemokine receptor 4 leads to the down-regulation of cell-to-cell contact, enhanced motility and proliferation in breast carcinoma cells. Cancer Res. 2006, 66, 5665–5675. [Google Scholar] [CrossRef] [PubMed]
- Devine, S.M.; Flomenberg, N.; Vesole, D.H.; Liesveld, J.; Weisdorf, D.; Badel, K.; Calandra, G.; DiPersio, J.F. Rapid mobilization of cd34+ cells following administration of the cxcr4 antagonist amd3100 to patients with multiple myeloma and non-hodgkin’s lymphoma. J. Clin. Oncol. 2004, 22, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Flomenberg, N.; Devine, S.M.; Dipersio, J.F.; Liesveld, J.L.; McCarty, J.M.; Rowley, S.D.; Vesole, D.H.; Badel, K.; Calandra, G. The use of amd3100 plus g-csf for autologous hematopoietic progenitor cell mobilization is superior to g-csf alone. Blood 2005, 106, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Scala, S. Molecular pathways: Targeting the cxcr4-cxcl12 axis--untapped potential in the tumor microenvironment. Clin. Cancer Res. 2015, 21, 4278–4285. [Google Scholar] [CrossRef]
- Peng, S.B.; Zhang, X.; Paul, D.; Kays, L.M.; Gough, W.; Stewart, J.; Uhlik, M.T.; Chen, Q.; Hui, Y.H.; Zamek-Gliszczynski, M.J.; et al. Identification of ly2510924, a novel cyclic peptide cxcr4 antagonist that exhibits antitumor activities in solid tumor and breast cancer metastatic models. Mol. Cancer Ther. 2015, 14, 480–490. [Google Scholar] [CrossRef]
- Peng, S.B.; Zhang, X.; Paul, D.; Kays, L.M.; Ye, M.; Vaillancourt, P.; Dowless, M.; Stancato, L.F.; Stewart, J.; Uhlik, M.T.; et al. Inhibition of cxcr4 by ly2624587, a fully humanized anti-cxcr4 antibody induces apoptosis of hematologic malignancies. PLoS ONE 2016, 11, e0150585. [Google Scholar] [CrossRef]
- Galsky, M.D.; Vogelzang, N.J.; Conkling, P.; Raddad, E.; Polzer, J.; Roberson, S.; Stille, J.R.; Saleh, M.; Thornton, D. A phase i trial of ly2510924, a cxcr4 peptide antagonist, in patients with advanced cancer. Clin. Cancer Res. 2014, 20, 3581–3588. [Google Scholar] [CrossRef]
- Arakaki, R.; Tamamura, H.; Premanathan, M.; Kanbara, K.; Ramanan, S.; Mochizuki, K.; Baba, M.; Fujii, N.; Nakashima, H. T134, a small-molecule cxcr4 inhibitor, has no cross-drug resistance with amd3100, a cxcr4 antagonist with a different structure. J. Virol. 1999, 73, 1719–1723. [Google Scholar] [PubMed]
- Torsvik, A.; Bjerkvig, R. Mesenchymal stem cell signaling in cancer progression. Cancer Treat. Rev. 2013, 39, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Quante, M.; Tu, S.P.; Tomita, H.; Gonda, T.; Wang, S.S.; Takashi, S.; Baik, G.H.; Shibata, W.; Diprete, B.; Betz, K.S.; et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 2011, 19, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Kato, I.; Niwa, A.; Heike, T.; Fujino, H.; Saito, M.K.; Umeda, K.; Hiramatsu, H.; Ito, M.; Morita, M.; Nishinaka, Y.; et al. Identification of hepatic niche harboring human acute lymphoblastic leukemic cells via the sdf-1/cxcr4 axis. PLoS ONE 2011, 6, e27042. [Google Scholar] [CrossRef] [PubMed]
- Roato, I.; Ferracini, R. Cancer stem cells, bone and tumor microenvironment: Key players in bone metastases. Cancers 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Farkas, D.; Kraskauskas, D.; Drake, J.I.; Alhussaini, A.A.; Kraskauskiene, V.; Bogaard, H.J.; Cool, C.D.; Voelkel, N.F.; Farkas, L. Cxcr4 inhibition ameliorates severe obliterative pulmonary hypertension and accumulation of c-kit(+) cells in rats. PLoS ONE 2014, 9, e89810. [Google Scholar] [CrossRef] [PubMed]
- Favre, S.; Gambini, E.; Nigro, P.; Scopece, A.; Bianciardi, P.; Caretti, A.; Pompilio, G.; Corno, A.F.; Vassalli, G.; von Segesser, L.K.; et al. Sildenafil attenuates hypoxic pulmonary remodelling by inhibiting bone marrow progenitor cells. J. Cell. Mol. Med. 2017, 21, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wu, P.; Huang, F.; Xu, M.; Chen, M.; Huang, K.; Li, G.P.; Xu, M.; Yao, D.; Wang, L. Baicalin attenuates chronic hypoxia-induced pulmonary hypertension via adenosine a2a receptor-induced sdf-1/cxcr4/pi3k/akt signaling. J. Biomed. Sci. 2017, 24, 52. [Google Scholar] [CrossRef]
- Yu, L.; Hales, C.A. Effect of chemokine receptor cxcr4 on hypoxia-induced pulmonary hypertension and vascular remodeling in rats. Respir. Res. 2011, 12, 21. [Google Scholar] [CrossRef]
- Drummond, S.; Ramachandran, S.; Torres, E.; Huang, J.; Hehre, D.; Suguihara, C.; Young, K.C. Cxcr4 blockade attenuates hyperoxia-induced lung injury in neonatal rats. Neonatology 2015, 107, 304–311. [Google Scholar] [CrossRef]
- Young, K.C.; Torres, E.; Hatzistergos, K.E.; Hehre, D.; Suguihara, C.; Hare, J.M. Inhibition of the sdf-1/cxcr4 axis attenuates neonatal hypoxia-induced pulmonary hypertension. Circ. Res. 2009, 104, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Gambaryan, N.; Perros, F.; Montani, D.; Cohen-Kaminsky, S.; Mazmanian, M.; Renaud, J.F.; Simonneau, G.; Lombet, A.; Humbert, M. Targeting of c-kit+ haematopoietic progenitor cells prevents hypoxic pulmonary hypertension. Eur. Respir. J. 2011, 37, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Zhang, N.; Wang, H.W.; Gao, P.; Yang, Q.P.; Wen, Q.P. Cxcr4 receptor overexpression in mesenchymal stem cells facilitates treatment of acute lung injury in rats. J. Biol. Chem. 2015, 290, 1994–2006. [Google Scholar] [CrossRef] [PubMed]
- Sartina, E.; Suguihara, C.; Ramchandran, S.; Nwajei, P.; Rodriguez, M.; Torres, E.; Hehre, D.; Devia, C.; Walters, M.J.; Penfold, M.E.; et al. Antagonism of cxcr7 attenuates chronic hypoxia-induced pulmonary hypertension. Pediatr. Res. 2012, 71, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, H.; van Ingen Schenau, D.S.; Bakker, N.E.; van Disseldorp, A.J.; Strik, A.; Hermens, L.S.; Koenen, T.B.; Krajnc-Franken, M.A.; Gossen, J.A. Early postnatal lethality and cardiovascular defects in cxcr7-deficient mice. Genesis 2008, 46, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto-Kataoka, T.; Hosen, N.; Sonobe, T.; Arita, Y.; Yasui, T.; Masaki, T.; Minami, M.; Inagaki, T.; Miyagawa, S.; Sawa, Y.; et al. Interleukin-6/interleukin-21 signaling axis is critical in the pathogenesis of pulmonary arterial hypertension. Proc. Natl. Acad. Sci. USA 2015, 112, E2677–E2686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kawaguchi, N.; Hayama, E.; Furutani, Y.; Nakanishi, T. High expression of cxcr4 and stem cell markers in a monocrotaline and chronic hypoxia-induced rat model of pulmonary arterial hypertension. Exp. Ther. Med. 2018, 15, 4615–4622. [Google Scholar] [CrossRef]
- Roe, N.D.; Xu, X.; Kandadi, M.R.; Hu, N.; Pang, J.; Weiser-Evans, M.C.; Ren, J. Targeted deletion of pten in cardiomyocytes renders cardiac contractile dysfunction through interruption of pink1-ampk signaling and autophagy. Biochim. Biophys. Acta 2015, 290–298. [Google Scholar] [CrossRef]
- Surai, P.F. Silymarin as a natural antioxidant: An overview of the current evidence and perspectives. Antioxidants 2015, 4, 204–247. [Google Scholar] [CrossRef]
- Saller, R.; Meier, R.; Brignoli, R. The use of silymarin in the treatment of liver diseases. Drugs 2001, 61, 2035–2063. [Google Scholar] [CrossRef]
- Singh, R.P.; Agarwal, R. Prostate cancer chemoprevention by silibinin: Bench to bedside. Mol. Carcinog. 2006, 45, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Crocenzi, F.A.; Roma, M.G. Silymarin as a new hepatoprotective agent in experimental cholestasis: New possibilities for an ancient medication. Curr. Med. Chem. 2006, 13, 1055–1074. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Agarwal, C.; Ichikawa, H.; Singh, R.P.; Aggarwal, B.B. Anticancer potential of silymarin: From bench to bed side. Anticancer Res. 2006, 26, 4457–4498. [Google Scholar] [PubMed]
- Kaur, M.; Agarwal, R. Silymarin and epithelial cancer chemoprevention: How close we are to bedside? Toxicol. Appl. Pharmacol. 2007, 224, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Agarwal, R. Mechanisms of action of novel agents for prostate cancer chemoprevention. Endocr. Relat. Cancer 2006, 13, 751–778. [Google Scholar] [CrossRef] [PubMed]
- Gazak, R.; Walterova, D.; Kren, V. Silybin and silymarin--new and emerging applications in medicine. Curr. Med. Chem. 2007, 14, 315–338. [Google Scholar] [CrossRef] [PubMed]
- Raina, K.; Agarwal, R. Combinatorial strategies for cancer eradication by silibinin and cytotoxic agents: Efficacy and mechanisms. Acta Pharmacol. Sin. 2007, 28, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Kroll, D.J.; Shaw, H.S.; Oberlies, N.H. Milk thistle nomenclature: Why it matters in cancer research and pharmacokinetic studies. Integr. Cancer Ther. 2007, 6, 110–119. [Google Scholar] [CrossRef]
- Abenavoli, L.; Capasso, R.; Milic, N.; Capasso, F. Milk thistle in liver diseases: Past, present, future. Phytother. Res. 2010, 24, 1423–1432. [Google Scholar] [CrossRef]
- Loguercio, C.; Festi, D. Silybin and the liver: From basic research to clinical practice. World J. Gastroenterol. 2011, 17, 2288–2301. [Google Scholar] [CrossRef]
- Ting, H.; Deep, G.; Agarwal, C.; Agarwal, R. The strategies to control prostate cancer by chemoprevention approaches. Mutat. Res. 2014, 760, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, D.; Vavrikova, E.; Cvak, L.; Kren, V. Chemistry of silybin. Nat. Prod. Rep. 2014, 31, 1138–1157. [Google Scholar] [CrossRef] [PubMed]
- Kazazis, C.E.; Evangelopoulos, A.A.; Kollas, A.; Vallianou, N.G. The therapeutic potential of milk thistle in diabetes. Rev. Diabet. Stud. 2014, 11, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Dhanalakshmi, S.; Singh, R.P.; Agarwal, C.; Agarwal, R. Silibinin inhibits constitutive and tnfalpha-induced activation of nf-kappab and sensitizes human prostate carcinoma du145 cells to tnfalpha-induced apoptosis. Oncogene 2002, 21, 1759–1767. [Google Scholar] [CrossRef]
- Singh, R.P.; Agarwal, R. A cancer chemopreventive agent silibinin, targets mitogenic and survival signaling in prostate cancer. Mutat. Res. 2004, 555, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, W.C.; Pan, W.L.; Law, W.K.; Hu, J.S.; Ip, D.T.; Waye, M.M.; Ng, T.B.; Wan, D.C. Silibinin, a novel chemokine receptor type 4 antagonist, inhibits chemokine ligand 12-induced migration in breast cancer cells. Phytomedicine 2014, 21, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.W.; Zhang, C.; Hong, T.; Liu, D.H.; Wang, C.; Li, J.; He, X.K.; Xu, W.D. Silibinin alleviates inflammation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes and has a therapeutic effect on arthritis in rats. Sci. Rep. 2018, 8, 3241. [Google Scholar] [CrossRef] [PubMed]
- Toxicology and carcinogenesis studies of milk thistle extract (cas no. 84604-20-6) in f344/n rats and b6c3f1 mice (feed studies). Natl. Toxicol. Program Tech. Rep. Ser. 2011, 565, 1–177.
- Hernandez, P.A.; Gorlin, R.J.; Lukens, J.N.; Taniuchi, S.; Bohinjec, J.; Francois, F.; Klotman, M.E.; Diaz, G.A. Mutations in the chemokine receptor gene cxcr4 are associated with whim syndrome, a combined immunodeficiency disease. Nat. Genet. 2003, 34, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. Cxcr4 signaling in health and disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Bachelerie, F. Cxcl12/cxcr4-axis dysfunctions: Markers of the rare immunodeficiency disorder whim syndrome. Dis. Markers 2010, 29, 189–198. [Google Scholar] [CrossRef] [PubMed]
- McDermott, D.H.; Pastrana, D.V.; Calvo, K.R.; Pittaluga, S.; Velez, D.; Cho, E.; Liu, Q.; Trout, H.H., 3rd; Neves, J.F.; Gardner, P.J.; et al. Plerixafor for the treatment of WHIM syndrome. N. Engl. J. Med. 2019, 380, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, M.; Moriuchi, H.; Margolis, D.M.; Fauci, A.S. Usf/c-myc enhances, while yin-yang 1 suppresses, the promoter activity of cxcr4, a coreceptor for hiv-1 entry. J. Immunol. 1999, 162, 5986–5992. [Google Scholar] [PubMed]
- Oldham, W.M.; Van Eps, N.; Preininger, A.M.; Hubbell, W.L.; Hamm, H.E. Mechanism of the receptor-catalyzed activation of heterotrimeric g proteins. Nat. Struct. Mol. Biol. 2006, 13, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.E.; Hatch, M.M.; Wu, N.; Muawad, S.A.; Hughes, C.C. Mtorc2 mediates cxcl12-induced angiogenesis. Angiogenesis 2016, 19, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Xia, Y.; Zuo, K.; Wang, Y.; Zhang, S.; Kuang, D.; Duan, Y.; Zhao, X.; Wang, G. Crosstalk between sdf-1/cxcr4 and sdf-1/cxcr7 in cardiac stem cell migration. Sci. Rep. 2015, 5, 16813. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, Z.; Wang, H.; Zhan, Y.; Li, G.; Yang, H.; Fei, Z.; Xu, Y.; Li, W. CXCR3 expression in colorectal cancer cells enhanced invasion through preventing CXCR4 internalization. Exp. Cell Res. 2018, 371, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.Y.; Wang, F.; Cui, S.X.; Gao, Z.H.; Qu, X.J. CXCR7/CXCR4 heterodimer-induced histone demethylation: A new mechanism of colorectal tumorigenesis. Oncogene 2018, 38, 1560–1575. [Google Scholar] [CrossRef]
- Spaks, A. Role of cxc group chemokines in lung cancer development and progression. J. Thorac. Dis. 2017, 9, S164–S171. [Google Scholar] [CrossRef]
- Saba, N.F.; Wang, Y.; Fu, H.; Koenig, L.; Khuri, F.R.; Shin, D.M.; Chen, Z.G. Association of cytoplasmic cxcr4 with loss of epithelial marker and activation of erk1/2 and akt signaling pathways in non-small-cell lung cancer. Clin. Lung Cancer 2017, 18, e203–e210. [Google Scholar] [CrossRef]
- Forte, E.; Chimenti, I.; Rosa, P.; Angelini, F.; Pagano, F.; Calogero, A.; Giacomello, A.; Messina, E. Emt/met at the crossroad of stemness, regeneration and oncogenesis: The ying-yang equilibrium recapitulated in cell spheroids. Cancers 2017, 9. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, N.; Zhang, T.-T.; Nakanishi, T. Involvement of CXCR4 in Normal and Abnormal Development. Cells 2019, 8, 185. https://doi.org/10.3390/cells8020185
Kawaguchi N, Zhang T-T, Nakanishi T. Involvement of CXCR4 in Normal and Abnormal Development. Cells. 2019; 8(2):185. https://doi.org/10.3390/cells8020185
Chicago/Turabian StyleKawaguchi, Nanako, Ting-Ting Zhang, and Toshio Nakanishi. 2019. "Involvement of CXCR4 in Normal and Abnormal Development" Cells 8, no. 2: 185. https://doi.org/10.3390/cells8020185



