The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy
Abstract
:1. Introduction—Protein Quality Control Mechanisms
2. Ub and Ubiquitination at the Crossroads of the UPS and Autophagy
3. Ub-Binding Proteins Functioning in the UPS
3.1. Rad23
3.2. Dsk2 and Ubiquilin Protein Families
3.3. Ddi1-Like Proteins
4. Ub-Binding Receptors in Autophagy
4.1. CUET Proteins
4.2. SQSTM1
4.3. NBR1
4.4. Plant NBR1-Like Proteins
4.5. OPTN
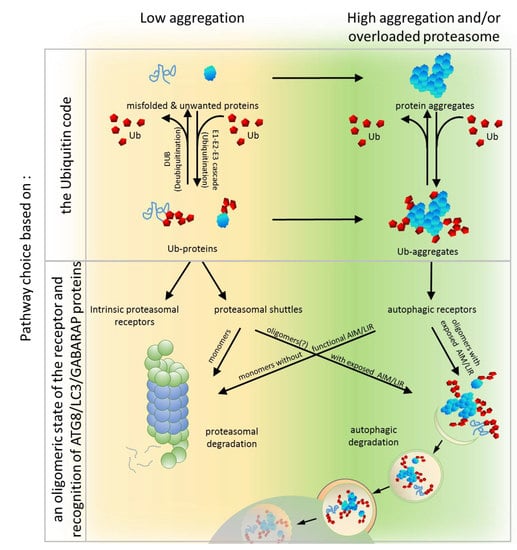
5. PQC Pathway Choice Based on the Ub Code
6. PQC Pathway Choice Based on an Oligomeric State of the Receptor and Recognition of ATG8/LC3/GABARAP Proteins
7. PQC Pathway Choice Based on Other Factors
7.1. Chaperone Assistance in the Pathway Choice
7.2. Conformational Changes of the Ub-Binding Receptors—The Intramolecular and Intermolecular Interactions
7.3. Post-Translational Modifications of the Ub-Binding Receptors
8. Suggested Model and Concluding Remarks
9. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Tanaka, K.; Matsuda, N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta 2014, 1843, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef]
- da Fonseca, P.C.; Morris, E.P. Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 2008, 283, 23305–23314. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Bedford, L.; Paine, S.; Sheppard, P.W.; Mayer, R.J.; Roelofs, J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010, 20, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Su, H.; Ranek, M.J. Protein quality control and degradation in cardiomyocytes. J. Mol. Cell. Cardiol. 2008, 45, 11–27. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, L.G.; Lindsten, K.; Masucci, M.G.; Dantuma, N.P. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 2002, 11, 2689–2700. [Google Scholar] [CrossRef] [Green Version]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- Taylor, J.P.; Tanaka, F.; Robitschek, J.; Sandoval, C.M.; Taye, A.; Markovic-Plese, S.; Fischbeck, K.H. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 2003, 12, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Suraweera, A.; Munch, C.; Hanssum, A.; Bertolotti, A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 2012, 48, 242–253. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef]
- Sugawara, K.; Suzuki, N.N.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 2004, 9, 611–618. [Google Scholar] [CrossRef]
- Schwarten, M.; Stoldt, M.; Mohrluder, J.; Willbold, D. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem. Biophys. Res. Commun. 2010, 395, 426–431. [Google Scholar] [CrossRef]
- Paz, Y.; Elazar, Z.; Fass, D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 2000, 275, 25445–25450. [Google Scholar] [CrossRef]
- Kumeta, H.; Watanabe, M.; Nakatogawa, H.; Yamaguchi, M.; Ogura, K.; Adachi, W.; Fujioka, Y.; Noda, N.N.; Ohsumi, Y.; Inagaki, F. The NMR structure of the autophagy-related protein Atg8. J. Biomol. NMR 2010, 47, 237–241. [Google Scholar] [CrossRef]
- Kouno, T.; Mizuguchi, M.; Tanida, I.; Ueno, T.; Kanematsu, T.; Mori, Y.; Shinoda, H.; Hirata, M.; Kominami, E.; Kawano, K. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 2005, 280, 24610–24617. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, X.; Teng, Y.B.; Hu, H.X.; Li, W.F. Structure of autophagy-related protein Atg8 from the silkworm Bombyx mori. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 787–790. [Google Scholar] [CrossRef]
- Coyle, J.E.; Qamar, S.; Rajashankar, K.R.; Nikolov, D.B. Structure of GABARAP in two conformations: Implications for GABA(A) receptor localization and tubulin binding. Neuron 2002, 33, 63–74. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural biology of the core autophagy machinery. Curr. Opin. Struct. Biol. 2017, 43, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Behrends, C.; Fulda, S. Receptor proteins in selective autophagy. Int. J. Cell Biol. 2012, 2012, 673290. [Google Scholar] [CrossRef]
- Lamark, T.; Johansen, T. Aggrephagy: Selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012, 2012, 736905. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Wang, X.; Xing, R.; Liu, K.; Gan, Q.; Tang, C.; Gao, Z.; Jian, Y.; Luo, S.; et al. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J. Cell Biol. 2017, 216, 1301–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Brown, A.I.; Rutenberg, A.D. Cluster coarsening on drops exhibits strong and sudden size-selectivity. Soft Matter 2015, 11, 3786–3793. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; Lamark, T.; Johansen, T.; Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009, 5, 732–733. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjorkoy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef]
- Lamark, T.; Kirkin, V.; Dikic, I.; Johansen, T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009, 8, 1986–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korac, J.; Schaeffer, V.; Kovacevic, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell. Sci. 2013, 126, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.C.; Li, H.Y.; Chen, G.C.; Chern, Y.; Tu, P.H. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 2015, 11, 685–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kravtsova-Ivantsiv, Y.; Sommer, T.; Ciechanover, A. The lysine48-based polyubiquitin chain proteasomal signal: Not a single child anymore. Angew. Chem. Int. Ed. Engl. 2013, 52, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade - an interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.; Walker, C.L. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell Cycle 2010, 9, 3685–3686. [Google Scholar] [CrossRef]
- Ji, C.H.; Kwon, Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Z.H. Coordination of autophagy with other cellular activities. Acta Pharmacol. Sin. 2013, 34, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X. The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim. Biophys. Acta 2015, 1852, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Lilienbaum, A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013, 4, 1–26. [Google Scholar] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Rajalingam, K.; Dikic, I. SnapShot: Expanding the ubiquitin code. Cell 2016, 164. [Google Scholar] [CrossRef] [PubMed]
- Woelk, T.; Sigismund, S.; Penengo, L.; Polo, S. The ubiquitination code: A signalling problem. Cell Div. 2007, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Madura, K. Rad23 and Rpn10: Perennial wallflowers join the melee. Trends Biochem. Sci. 2004, 29, 637–640. [Google Scholar] [CrossRef]
- Clarke, D.J.; Mondesert, G.; Segal, M.; Bertolaet, B.L.; Jensen, S.; Wolff, M.; Henze, M.; Reed, S.I. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 2001, 21, 1997–2007. [Google Scholar] [CrossRef]
- Elsasser, S.; Chandler-Militello, D.; Muller, B.; Hanna, J.; Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004, 279, 26817–26822. [Google Scholar] [CrossRef]
- Gomez, T.A.; Kolawa, N.; Gee, M.; Sweredoski, M.J.; Deshaies, R.J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011, 9, 33. [Google Scholar] [CrossRef]
- Hartmann-Petersen, R.; Gordon, C. Integral UBL domain proteins: A family of proteasome interacting proteins. Semin. Cell Dev. Biol. 2004, 15, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Schubert, H.L.; Hill, C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005, 6, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplun, L.; Tzirkin, R.; Bakhrat, A.; Shabek, N.; Ivantsiv, Y.; Raveh, D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of HO endonuclease. Mol. Cell. Biol. 2005, 25, 5355–5362. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Shih, A.H.; Zhou, P.; Kumar, S.; Soccio, R.E.; Kedersha, N.L.; Gill, G.; Howley, P.M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 2000, 6, 409–419. [Google Scholar] [CrossRef]
- Lambertson, D.; Chen, L.; Madura, K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 1999, 153, 69–79. [Google Scholar] [PubMed]
- Saeki, Y.; Saitoh, A.; Toh-e, A.; Yokosawa, H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2002, 293, 986–992. [Google Scholar] [CrossRef]
- Elsasser, S.; Finley, D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005, 7, 742–749. [Google Scholar] [CrossRef]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef]
- Walters, K.J.; Kleijnen, M.F.; Goh, A.M.; Wagner, G.; Howley, P.M. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002, 41, 1767–1777. [Google Scholar] [CrossRef]
- Mueller, T.D.; Feigon, J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 2002, 319, 1243–1255. [Google Scholar] [CrossRef]
- Dieckmann, T.; Withers-Ward, E.S.; Jarosinski, M.A.; Liu, C.F.; Chen, I.S.; Feigon, J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat. Struct. Biol. 1998, 5, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Mueller, T.D.; Kamionka, M.; Feigon, J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 2004, 279, 11926–11936. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Gabriely, G.; Kama, R.; Gelin-Licht, R.; Gerst, J.E. Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol. Biol. Cell 2008, 19, 3625–3637. [Google Scholar] [CrossRef]
- Sirkis, R.; Gerst, J.E.; Fass, D. Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 2006, 364, 376–387. [Google Scholar] [CrossRef]
- Watkins, J.F.; Sung, P.; Prakash, L.; Prakash, S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 1993, 13, 7757–7765. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Heinen, C.; Hoogstraten, D. The ubiquitin receptor Rad23: At the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009, 8, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Muller, B.; Feng, M.T.; Tubing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y.; Sone, T.; Toh-e, A.; Yokosawa, H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002, 296, 813–819. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Osmulski, P.A.; Gaczynska, M.; Glickman, M.H. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008, 15, 573–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, K.J.; Lech, P.J.; Goh, A.M.; Wang, Q.; Howley, P.M. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc. Natl. Acad. Sci. USA 2003, 100, 12694–12699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, K.S.; Lee, K.J.; Bae, S.H.; Kim, B.K.; Kim, K.A.; Choi, B.S. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J. Biol. Chem. 2003, 278, 36621–36627. [Google Scholar] [CrossRef] [PubMed]
- Raasi, S.; Varadan, R.; Fushman, D.; Pickart, C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005, 12, 708–714. [Google Scholar] [CrossRef]
- Rao, H.; Sastry, A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002, 277, 11691–11695. [Google Scholar] [CrossRef]
- Chen, L.; Shinde, U.; Ortolan, T.G.; Madura, K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001, 2, 933–938. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Madura, K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002, 22, 4902–4913. [Google Scholar] [CrossRef]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001, 8, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Ortolan, T.G.; Tongaonkar, P.; Lambertson, D.; Chen, L.; Schauber, C.; Madura, K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2000, 2, 601–608. [Google Scholar] [CrossRef]
- Raasi, S.; Pickart, C.M. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 2003, 278, 8951–8959. [Google Scholar] [CrossRef] [PubMed]
- Biggins, S.; Ivanovska, I.; Rose, M.D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1996, 133, 1331–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, C.R.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Madura, K. Evidence for distinct functions for human DNA repair factors hHR23A and hHR23B. FEBS Lett. 2006, 580, 3401–3408. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, M.; Sasaki, T.; Nishimoto, T.; Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 2002, 99, 745–750. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Funakoshi, M.; Inoue, K.; Kobayashi, H. Identification of two isoforms of Dsk2-related protein XDRP1 in Xenopus eggs. Biochem. Biophys. Res. Commun. 2006, 350, 768–773. [Google Scholar] [CrossRef]
- Matiuhin, Y.; Kirkpatrick, D.S.; Ziv, I.; Kim, W.; Dakshinamurthy, A.; Kleifeld, O.; Gygi, S.P.; Reis, N.; Glickman, M.H. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 2008, 32, 415–425. [Google Scholar] [CrossRef]
- Lu, K.; den Brave, F.; Jentsch, S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 2017, 19, 732–739. [Google Scholar] [CrossRef]
- Zhao, G.; Zhou, X.; Wang, L.; Li, G.; Kisker, C.; Lennarz, W.J.; Schindelin, H. Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways. J. Biol. Chem. 2006, 281, 13751–13761. [Google Scholar] [CrossRef] [PubMed]
- El Ayadi, A.; Stieren, E.S.; Barral, J.M.; Boehning, D. Ubiquilin-1 and protein quality control in Alzheimer disease. Prion 2013, 7, 164–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Su, V.; Kurata, W.E.; Jin, C.; Lau, A.F. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J. Biol. Chem. 2008, 283, 5748–5759. [Google Scholar] [CrossRef] [PubMed]
- Heir, R.; Ablasou, C.; Dumontier, E.; Elliott, M.; Fagotto-Kaufmann, C.; Bedford, F.K. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006, 7, 1252–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothenberg, C.; Srinivasan, D.; Mah, L.; Kaushik, S.; Peterhoff, C.M.; Ugolino, J.; Fang, S.; Cuervo, A.M.; Nixon, R.A.; Monteiro, M.J. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 2010, 19, 3219–3232. [Google Scholar] [CrossRef] [Green Version]
- N’Diaye, E.N.; Kajihara, K.K.; Hsieh, I.; Morisaki, H.; Debnath, J.; Brown, E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009, 10, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Brown, E.J. Ubiquilins in the crosstalk among proteolytic pathways. Biol. Chem. 2012, 393, 441–447. [Google Scholar] [CrossRef]
- Ivantsiv, Y.; Kaplun, L.; Tzirkin-Goldin, R.; Shabek, N.; Raveh, D. Unique role for the UbL-UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol. Cell. Biol. 2006, 26, 1579–1588. [Google Scholar] [CrossRef]
- Voloshin, O.; Bakhrat, A.; Herrmann, S.; Raveh, D. Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS ONE 2012, 7, e39210. [Google Scholar] [CrossRef]
- Kaplun, L.; Ivantsiv, Y.; Kornitzer, D.; Raveh, D. Functions of the DNA damage response pathway target HO endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 2000, 97, 10077–10082. [Google Scholar] [CrossRef]
- Zhang, D.; Raasi, S.; Fushman, D. Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 2008, 377, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, U.; Zhang, D.; Walker, O.; Krutauz, D.; Castaneda, C.A.; Chaturvedi, A.; Chen, T.Y.; Reis, N.; Glickman, M.H.; Fushman, D. DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure 2015, 23, 542–557. [Google Scholar] [CrossRef]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J. Mol. Biol. 2001, 313, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Siva, M.; Svoboda, M.; Veverka, V.; Trempe, J.F.; Hofmann, K.; Kozisek, M.; Hexnerova, R.; Sedlak, F.; Belza, J.; Brynda, J.; et al. Human DNA-damage-inducible 2 protein is structurally and functionally distinct from its yeast ortholog. Sci. Rep. 2016, 6, 30443. [Google Scholar] [CrossRef]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Marchbank, K.; Solomon, E.; Whitehouse, C.; Gautel, M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009, 583, 1846–1852. [Google Scholar] [CrossRef] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, R.; Tanaka, K.; Komatsu, M. Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett. 2014, 588, 822–828. [Google Scholar] [CrossRef] [Green Version]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell. Sci. 2013, 126, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Isakson, P.; Lystad, A.H.; Breen, K.; Koster, G.; Stenmark, H.; Simonsen, A. TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy 2013, 9, 1955–1964. [Google Scholar] [CrossRef] [Green Version]
- Narendra, D.; Kane, L.A.; Hauser, D.N.; Fearnley, I.M.; Youle, R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 2010, 6, 1090–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Verlhac, P.; Gregoire, I.P.; Azocar, O.; Petkova, D.S.; Baguet, J.; Viret, C.; Faure, M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host. Microbe 2015, 17, 515–525. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- von Muhlinen, N.; Thurston, T.; Ryzhakov, G.; Bloor, S.; Randow, F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy 2010, 6, 288–289. [Google Scholar] [CrossRef]
- Mitra, S.; Traughber, C.A.; Brannon, M.K.; Gomez, S.; Capelluto, D.G. Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides. J. Biol. Chem. 2013, 288, 25780–25791. [Google Scholar] [CrossRef]
- Ankem, G.; Mitra, S.; Sun, F.; Moreno, A.C.; Chutvirasakul, B.; Azurmendi, H.F.; Li, L.; Capelluto, D.G. The C2 domain of Tollip, a Toll-like receptor signalling regulator, exhibits broad preference for phosphoinositides. Biochem. J. 2011, 435, 597–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capelluto, D.G. Tollip: A multitasking protein in innate immunity and protein trafficking. Microbes Infect. 2012, 14, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Visvikis, O.; Boyer, L.; Torrino, S.; Doye, A.; Lemonnier, M.; Lores, P.; Rolando, M.; Flatau, G.; Mettouchi, A.; Bouvard, D.; et al. Escherichia coli producing CNF1 toxin hijacks Tollip to trigger Rac1-dependent cell invasion. Traffic 2011, 12, 579–590. [Google Scholar] [CrossRef]
- Doi, H.; Mitsui, K.; Kurosawa, M.; Machida, Y.; Kuroiwa, Y.; Nukina, N. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004, 571, 171–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oguro, A.; Kubota, H.; Shimizu, M.; Ishiura, S.; Atomi, Y. Protective role of the ubiquitin binding protein Tollip against the toxicity of polyglutamine-expansion proteins. Neurosci. Lett. 2011, 503, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Bjorkoy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankiv, S.; Lamark, T.; Bruun, J.A.; Overvatn, A.; Bjorkoy, G.; Johansen, T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010, 285, 5941–5953. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef]
- Lin, X.; Li, S.; Zhao, Y.; Ma, X.; Zhang, K.; He, X.; Wang, Z. Interaction domains of p62: A bridge between p62 and selective autophagy. DNA Cell Biol. 2013, 32, 220–227. [Google Scholar] [CrossRef]
- Seibenhener, M.L.; Babu, J.R.; Geetha, T.; Wong, H.C.; Krishna, N.R.; Wooten, M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004, 24, 8055–8068. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.; Seibenhener, M.L.; Chen, L.; Madura, K.; Wooten, M.W. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 2008, 374, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, N.; Zhang, L.; Li, R.; Fu, W.; Ma, K.; Li, X.; Wang, L.; Wang, J.; Zhang, H.; et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol. Cell 2016, 63, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puvirajesinghe, T.M.; Bertucci, F.; Jain, A.; Scerbo, P.; Belotti, E.; Audebert, S.; Sebbagh, M.; Lopez, M.; Brech, A.; Finetti, P.; et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat. Commun. 2016, 7, 10318. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Qiang, L.; Sample, A.; Shah, P.; He, Y.Y. NF-kappaB signaling activation induced by chloroquine requires autophagosome, p62 protein, and c-Jun N-terminal Kinase (JNK) signaling and promotes tumor cell resistance. J. Biol. Chem. 2017, 292, 3379–3388. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.H.; Lamark, T.; Isakson, P.; Finley, K.; Larsen, K.B.; Brech, A.; Overvatn, A.; Stenmark, H.; Bjorkoy, G.; Simonsen, A.; et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010, 6, 330–344. [Google Scholar] [CrossRef] [Green Version]
- Itakura, E.; Mizushima, N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 2011, 192, 17–27. [Google Scholar] [CrossRef]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife 2015, 4, e08941. [Google Scholar] [CrossRef]
- Yan, J.; Seibenhener, M.L.; Calderilla-Barbosa, L.; Diaz-Meco, M.T.; Moscat, J.; Jiang, J.; Wooten, M.W.; Wooten, M.C. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS ONE 2013, 8, e76016. [Google Scholar] [CrossRef]
- Marchbank, K.; Waters, S.; Roberts, R.G.; Solomon, E.; Whitehouse, C.A. MAP1B Interaction with the FW domain of the autophagic receptor Nbr1 facilitates its association to the microtubule network. Int. J. Cell Biol. 2012, 2012, 208014. [Google Scholar] [CrossRef] [PubMed]
- Mardakheh, F.K.; Auciello, G.; Dafforn, T.R.; Rappoport, J.Z.; Heath, J.K. Nbr1 is a novel inhibitor of ligand-mediated receptor tyrosine kinase degradation. Mol. Cell. Biol. 2010, 30, 5672–5685. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Perander, M.; Outzen, H.; Kristiansen, K.; Overvatn, A.; Michaelsen, E.; Bjorkoy, G.; Johansen, T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003, 278, 34568–34581. [Google Scholar] [CrossRef] [PubMed]
- Christian, F.; Krause, E.; Houslay, M.D.; Baillie, G.S. PKA phosphorylation of p62/SQSTM1 regulates PB1 domain interaction partner binding. Biochim. Biophys. Acta 2014, 1843, 2765–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.I.; Gill, D.J.; Perisic, O.; Quinn, M.T.; Williams, R.L. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 2003, 12, 39–50. [Google Scholar] [CrossRef]
- Muller, S.; Kursula, I.; Zou, P.; Wilmanns, M. Crystal structure of the PB1 domain of NBR1. FEBS Lett. 2006, 580, 341–344. [Google Scholar] [CrossRef]
- Walinda, E.; Morimoto, D.; Sugase, K.; Konuma, T.; Tochio, H.; Shirakawa, M. Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin. J. Biol. Chem. 2014, 289, 13890–13902. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front. Plant Sci. 2014, 5, 13. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [Green Version]
- Kachaner, D.; Filipe, J.; Laplantine, E.; Bauch, A.; Bennett, K.L.; Superti-Furga, G.; Israel, A.; Weil, R. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Mol. Cell 2012, 45, 553–566. [Google Scholar] [CrossRef] [PubMed]
- del Toro, D.; Alberch, J.; Lazaro-Dieguez, F.; Martin-Ibanez, R.; Xifro, X.; Egea, G.; Canals, J.M. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell 2009, 20, 1478–1492. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wu, C.J.; Zhao, Y.; Ashwell, J.D. Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 2007, 17, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Anborgh, P.H.; Godin, C.; Pampillo, M.; Dhami, G.K.; Dale, L.B.; Cregan, S.P.; Truant, R.; Ferguson, S.S. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J. Biol. Chem. 2005, 280, 34840–34848. [Google Scholar] [CrossRef]
- Sahlender, D.A.; Roberts, R.C.; Arden, S.D.; Spudich, G.; Taylor, M.J.; Luzio, J.P.; Kendrick-Jones, J.; Buss, F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005, 169, 285–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattula, K.; Peranen, J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000, 10, 1603–1606. [Google Scholar] [CrossRef] [Green Version]
- Slowicka, K.; Vereecke, L.; van Loo, G. Cellular functions of Optineurin in health and disease. Trends Immunol. 2016, 37, 621–633. [Google Scholar] [CrossRef]
- Gleason, C.E.; Ordureau, A.; Gourlay, R.; Arthur, J.S.; Cohen, P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. J. Biol. Chem. 2011, 286, 35663–35674. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, P.; Gao, H.; Gu, Y.; Yang, J.; Peng, H.; Xu, X.; Wang, H.; Yang, M.; Liu, X.; et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 2014, 26, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, D.A.; Waxse, B.J.; Arden, S.D.; Bright, N.A.; Kendrick-Jones, J.; Buss, F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat. Cell Biol. 2012, 14, 1024–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, H.; Shen, X.; Park, B.; Yue, B.Y. Posttranslational modifications, localization, and protein interactions of optineurin, the product of a glaucoma gene. PLoS ONE 2010, 5, e9168. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Ying, H.; Qiu, Y.; Park, J.S.; Shyam, R.; Chi, Z.L.; Iwata, T.; Yue, B.Y. Processing of optineurin in neuronal cells. J. Biol. Chem. 2011, 286, 3618–3629. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.E.; Riley, B.E.; Shaler, T.A.; Trevino, R.S.; Becker, C.H.; Schulman, H.; Kopito, R.R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 8, 691–696. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Reiss, Y.; Fried, V.A.; Hershko, A.; Yoon, J.K.; Gonda, D.K.; Sangan, P.; Copeland, N.G.; Jenkins, N.A.; Varshavsky, A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7898–7903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasaki, T.; Mulder, L.C.; Iwamatsu, A.; Lee, M.J.; Davydov, I.V.; Varshavsky, A.; Muesing, M.; Kwon, Y.T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005, 25, 7120–7136. [Google Scholar] [CrossRef]
- Liu, W.; Shang, Y.; Li, W. gp78 elongates of polyubiquitin chains from the distal end through the cooperation of its G2BR and CUE domains. Sci. Rep. 2014, 4, 7138. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.C.; Huibregtse, J.M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 2009, 29, 3307–3318. [Google Scholar] [CrossRef]
- Suber, T.; Wei, J.; Jacko, A.M.; Nikolli, I.; Zhao, Y.; Zhao, J.; Mallampalli, R.K. SCF(FBXO17) E3 ligase modulates inflammation by regulating proteasomal degradation of glycogen synthase kinase-3beta in lung epithelia. J. Biol. Chem. 2017, 292, 7452–7461. [Google Scholar] [CrossRef]
- McKeon, J.E.; Sha, D.; Li, L.; Chin, L.S. Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell. Mol. Life Sci. 2015, 72, 1811–1824. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Prajapati, P.; Sripada, L.; Singh, K.; Singh, R.; Singh, A.K.; Singh, R. TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim. Biophys. Acta 2013, 1833, 3134–3144. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.V.; Soares, A.R.; Ramalho, J.S.; Pereira, P.; Girao, H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by chaperone-mediated autophagy. Sci. Rep. 2015, 5, 10210. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Stanhill, A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.M.; Wong, E.S.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef]
- Lin, Y.L.; Sung, S.C.; Tsai, H.L.; Yu, T.T.; Radjacommare, R.; Usharani, R.; Fatimababy, A.S.; Lin, H.Y.; Wang, Y.Y.; Fu, H. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 2011, 23, 2754–2773. [Google Scholar] [CrossRef]
- Farmer, L.M.; Book, A.J.; Lee, K.H.; Lin, Y.L.; Fu, H.; Vierstra, R.D. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 2010, 22, 124–142. [Google Scholar] [CrossRef]
- Wooten, M.W.; Geetha, T.; Babu, J.R.; Seibenhener, M.L.; Peng, J.; Cox, N.; Diaz-Meco, M.T.; Moscat, J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 2008, 283, 6783–6789. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Gallagher, T.R.; Cavey, J.R.; Sheppard, P.W.; Ralston, S.H.; Layfield, R.; Searle, M.S. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J. Biol. Chem. 2008, 283, 5427–5440. [Google Scholar] [CrossRef]
- Riley, B.E.; Kaiser, S.E.; Shaler, T.A.; Ng, A.C.; Hara, T.; Hipp, M.S.; Lage, K.; Xavier, R.J.; Ryu, K.Y.; Taguchi, K.; et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: A potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 2010, 191, 537–552. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 163–181. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef]
- Matsumoto, M.L.; Wickliffe, K.E.; Dong, K.C.; Yu, C.; Bosanac, I.; Bustos, D.; Phu, L.; Kirkpatrick, D.S.; Hymowitz, S.G.; Rape, M.; et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 2010, 39, 477–484. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.P.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef]
- Kirkpatrick, D.S.; Hathaway, N.A.; Hanna, J.; Elsasser, S.; Rush, J.; Finley, D.; King, R.W.; Gygi, S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006, 8, 700–710. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Saeki, Y.; Kudo, T.; Sone, T.; Kikuchi, Y.; Yokosawa, H.; Toh-e, A.; Tanaka, K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009, 28, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, S.; Zheng, H. The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau). Autophagy 2018, 1–16. [Google Scholar] [CrossRef]
- Chuang, K.H.; Liang, F.; Higgins, R.; Wang, Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol. Biol. Cell 2016, 27, 2025–2036. [Google Scholar] [CrossRef] [Green Version]
- Meriin, A.B.; Zhang, X.; Miliaras, N.B.; Kazantsev, A.; Chernoff, Y.O.; McCaffery, J.M.; Wendland, B.; Sherman, M.Y. Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol. Cell. Biol. 2003, 23, 7554–7565. [Google Scholar] [CrossRef]
- Rothenberg, C.; Monteiro, M.J. Ubiquilin at a crossroads in protein degradation pathways. Autophagy 2010, 6, 979–980. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Arnott, D.; Brown, E.J. Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 2013, 14, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46.e37. [Google Scholar] [CrossRef]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.; Lamark, T.; et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef]
- Han, H.; Wei, W.; Duan, W.; Guo, Y.; Li, Y.; Wang, J.; Bi, Y.; Li, C. Autophagy-linked FYVE protein (Alfy) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). In Vitro Cell. Dev. Biol. Anim. 2015, 51, 249–263. [Google Scholar] [CrossRef]
- Rui, Y.N.; Xu, Z.; Patel, B.; Chen, Z.; Chen, D.; Tito, A.; David, G.; Sun, Y.; Stimming, E.F.; Bellen, H.J.; et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015, 17, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Ohtsubo, M.; Hotta, Y.; Minoshima, S. Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: Possible relationship with glaucoma pathology. PLoS ONE 2014, 9, e101206. [Google Scholar] [CrossRef]
- Elliott, E.; Tsvetkov, P.; Ginzburg, I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J. Biol. Chem. 2007, 282, 37276–37284. [Google Scholar] [CrossRef]
- Luders, J.; Demand, J.; Hohfeld, J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 2000, 275, 4613–4617. [Google Scholar] [CrossRef]
- Behl, C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Carra, S.; Seguin, S.J.; Landry, J. HspB8 and Bag3: A new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 2008, 4, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Carra, S.; Seguin, S.J.; Lambert, H.; Landry, J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 2008, 283, 1437–1444. [Google Scholar] [CrossRef]
- Minoia, M.; Boncoraglio, A.; Vinet, J.; Morelli, F.F.; Brunsting, J.F.; Poletti, A.; Krom, S.; Reits, E.; Kampinga, H.H.; Carra, S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 2014, 10, 1603–1621. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Kaya, A.M.; Wolfrum, U.; Clement, A.M.; Behl, C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011, 12, 149–156. [Google Scholar] [CrossRef] [Green Version]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Kessens, R.; Dickman, M.B. A plant Bcl-2-associated athanogene is proteolytically activated to confer fungal resistance. Microb. Cell 2016, 3, 224–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dickman, M. Processing of AtBAG6 triggers autophagy and fungal resistance. Plant Signal. Behav. 2016, 11, e1175699. [Google Scholar] [CrossRef]
- Sebti, S.; Prebois, C.; Perez-Gracia, E.; Bauvy, C.; Desmots, F.; Pirot, N.; Gongora, C.; Bach, A.S.; Hubberstey, A.V.; Palissot, V.; et al. BAG6/BAT3 modulates autophagy by affecting EP300/p300 intracellular localization. Autophagy 2014, 10, 1341–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, E.D.; Hasan, N.; Trempe, J.F.; Fonso, L.; Noble, M.E.; Endicott, J.A.; Johnson, L.N.; Brown, N.R. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 177–188. [Google Scholar] [CrossRef]
- Wang, Q.; Goh, A.M.; Howley, P.M.; Walters, K.J. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 2003, 42, 13529–13535. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Funakoshi, M.; Endicott, J.A.; Kobayashi, H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem. Biophys. Res. Commun. 2005, 336, 530–535. [Google Scholar] [CrossRef]
- Ford, D.L.; Monteiro, M.J. Dimerization of ubiquilin is dependent upon the central region of the protein: Evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J. 2006, 399, 397–404. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, N.; Koepp, D.M.; Walters, K.J. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J. Mol. Biol. 2007, 365, 1093–1101. [Google Scholar] [CrossRef]
- Hirano, Y.; Yoshinaga, S.; Ogura, K.; Yokochi, M.; Noda, Y.; Sumimoto, H.; Inagaki, F. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J. Biol. Chem. 2004, 279, 31883–31890. [Google Scholar] [CrossRef] [PubMed]
- Su, V.; Lau, A.F. Ubiquitin-like and ubiquitin-associated domain proteins: Significance in proteasomal degradation. Cell. Mol. Life Sci. 2009, 66, 2819–2833. [Google Scholar] [CrossRef]
- Isogai, S.; Morimoto, D.; Arita, K.; Unzai, S.; Tenno, T.; Hasegawa, J.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; Shirakawa, M.; et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 2011, 286, 31864–31874. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.A.; Sun, Y.; Jiang, Y.P.; Bott, A.J.; Jaber, N.; Dou, Z.; Yang, B.; Chen, J.S.; Catanzaro, J.M.; Du, C.; et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol. Cell 2016, 61, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.Y.; Chen, L.; Ko, B.T.; Shen, Y.H.; Li, Y.T.; Chen, B.R.; Lin, K.T.; Madura, K.; Chuang, S.M. Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J. Mol. Biol. 2014, 426, 4049–4060. [Google Scholar] [CrossRef]
- Godderz, D.; Giovannucci, T.A.; Lalakova, J.; Menendez-Benito, V.; Dantuma, N.P. The deubiquitylating enzyme Ubp12 regulates Rad23-dependent proteasomal degradation. J. Cell. Sci. 2017, 130, 3336–3346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ghosh, S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J. Biol. Chem. 2002, 277, 7059–7065. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, S.; Wu, H.; Gao, R.; Rao, G.; Wang, D.; Chen, Z.; Ma, B.; Wang, H.; Sui, N.; et al. Parkin promotes proteasomal degradation of p62: Implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell 2016, 7, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, G.; Wada, K.; Okuno, M.; Kurosawa, M.; Nukina, N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 2011, 44, 279–289. [Google Scholar] [CrossRef]
- Heath, R.J.; Goel, G.; Baxt, L.A.; Rush, J.S.; Mohanan, V.; Paulus, G.L.C.; Jani, V.; Lassen, K.G.; Xavier, R.J. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell. Rep. 2016, 17, 2183–2194. [Google Scholar] [CrossRef]
- Kalogeropulou, A.F.; Zhao, J.; Bolliger, M.F.; Memou, A.; Narasimha, S.; Molitor, T.P.; Wilson, W.H.; Rideout, H.J.; Nichols, R.J. P62/SQSTM1 is a novel leucine-rich repeat kinase 2 (LRRK2) substrate that enhances neuronal toxicity. Biochem. J. 2018, 475, 1271–1293. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Amanchy, R.; Greis, K.; Diaz-Meco, M.T.; Moscat, J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol. Cell. Biol. 2011, 31, 105–117. [Google Scholar] [CrossRef]
- Ha, S.; Jeong, S.H.; Yi, K.; Chung, K.M.; Hong, C.J.; Kim, S.W.; Kim, E.K.; Yu, S.W. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J. Biol. Chem. 2017, 292, 13795–13808. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsujimura, A.; Taguchi, K.; Tanaka, M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 2017, 13, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Simmons, A.N.; Kajino-Sakamoto, R.; Tsuji, Y.; Ninomiya-Tsuji, J. TAK1 regulates the Nrf2 antioxidant system through modulating p62/SQSTM1. Antioxid. Redox Signal. 2016, 25, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lachenmayer, M.L.; Wu, S.; Liu, W.; Kundu, M.; Wang, R.; Komatsu, M.; Oh, Y.J.; Zhao, Y.; Yue, Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015, 11, e1004987. [Google Scholar] [CrossRef] [PubMed]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef]
- Lee, Y.; Chou, T.F.; Pittman, S.K.; Keith, A.L.; Razani, B.; Weihl, C.C. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell. Rep. 2017, 19, 188–202. [Google Scholar] [CrossRef]
- Jongsma, M.L.; Berlin, I.; Wijdeven, R.H.; Janssen, L.; Janssen, G.M.; Garstka, M.A.; Janssen, H.; Mensink, M.; van Veelen, P.A.; Spaapen, R.M.; et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef]
- Lin, Q.; Dai, Q.; Meng, H.; Sun, A.; Wei, J.; Peng, K.; Childress, C.; Chen, M.; Shao, G.; Yang, W. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J. Cell. Sci. 2017, 130, 3839–3850. [Google Scholar] [CrossRef] [PubMed]
- Nicot, A.S.; Lo Verso, F.; Ratti, F.; Pilot-Storck, F.; Streichenberger, N.; Sandri, M.; Schaeffer, L.; Goillot, E. Phosphorylation of NBR1 by GSK3 modulates protein aggregation. Autophagy 2014, 10, 1036–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Chou, T.F.; Pittman, S.K.; Keith, A.L.; Razani, B.; Weihl, C.C. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell. Rep. 2017, 20, 1994. [Google Scholar] [CrossRef]
- Jacomin, A.C.; Samavedam, S.; Promponas, V.; Nezis, I.P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016, 12, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Birgisdottir, A.B.; Lamark, T.; Johansen, T. The LIR motif—Crucial for selective autophagy. J. Cell. Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Sung, K.S.; Hwang, J.; Kim, K.A.; Yu, J.E.; Yoo, Y.D.; Jang, J.M.; Han, D.H.; Molstad, M.; Kim, J.G.; et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015, 17, 917–929. [Google Scholar] [CrossRef] [Green Version]
- Isakson, P.; Holland, P.; Simonsen, A. The role of ALFY in selective autophagy. Cell Death Differ. 2013, 20, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Birkeland, H.C.; Gillooly, D.J.; Mizushima, N.; Kuma, A.; Yoshimori, T.; Slagsvold, T.; Brech, A.; Stenmark, H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell. Sci. 2004, 117, 4239–4251. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Species | Ub-Binding Protein | Modification/s | Function | Reference/s |
|---|---|---|---|---|
| Yeast | Rad23 | S47 and S73 phosphorylation | Inhibits binding of Rad23 to proteasome | [225] |
| UbL deubiquitination by Ubp12 | Stabilizes Rad23-substrate binding by inhibiting proteasomal degradation of substrate | [226] | ||
| Dsk2 | ubiquitination | Reduces capacity to bind poly-ubiquitinated proteins | ||
| Ddi1 | No data | |||
| Cue5 | No data | |||
| Human | HR23B | No data | ||
| Ubiquilin1-4 | No data | |||
| Ddi1 | No data | |||
| TOLLIP | Phosphorylation by IRAK1 | Regulates TLR-mediated cell activation by dissociation of IRAK1 from TOLLIP | [227] | |
| SQSTM1 | K7 ubiquitination by TRIM21 | Abrogates oligomerization via PB1 domain and inhibits sequestration activity | [224] | |
| K13 ubiquitination by Parkin | Promotes the proteasomal degradation of SQSTM1 | [228] | ||
| S24, S207, S403 phosphorylation | Role unknown, probably autophagy stimulation as a response to the MG132 treatment | [229] | ||
| K91 and K189 ubiquitination by RNF166 | Mediates xenophagy, K29- polyubiquitination and K33-polyubiquitination of SQSTM1 | [230] | ||
| T138 phosphorylation by LRRK | Role unknown, increases the neurotoxicity | [231] | ||
| T269 and S272 phosphorylation by CDK1 | Regulates exit from mitosis | [232] | ||
| S294 phosphorylation by AMPK | Induces mitophagy and autophagic cell death | [233] | ||
| S349 phosphorylation by CK1, TAK1 | Enhances the binding affinity between KEAP1 and SQSTM1, activates the KEAP1-NRF2 pathway | [234,235,236] | ||
| S403 phosphorylation by ULK1, CK2, TBK1 | Attenuates affinity to Ub | [229,237,238] | ||
| S407 phosphorylation by ULK1 | Attenuates affinity to Ub, facilitates phosphorylation of S405 | [233,234,237] | ||
| K420 ubiquitination by KEAP1/CUL3 | Increases sequestering activity and degradation | [233,234,237,239] | ||
| UBA domain ubiquitination by RNF26 | Facilitates vesicular cargo sorting | [239,240] | ||
| PB1 domain ubiquitination by NEDD4 | Facilitates inclusion body formation and autophagy mediated by SQSTM1 | [241] | ||
| NBR1 | T586 phosphorylation by GSK3 | Inhibits aggregation of ubiquitinated proteins and their degradation | [242] | |
| OPTN | S177 phosphorylation by TBK1 | Activates LIR motif and promotes LC3 binding | [243] | |
| S473 and S513 phosphorylation by TBK1 | Promotes poly-Ub binding | [243] | ||
| S473 phosphorylation | Promotes poly-Ub binding during mitophagy | [114] | ||
| S177 and S513 phosphorylation by IKKβ | Role unknown probably promoting autophagy | [158] | ||
| Plant | RAD23A-D | No data | ||
| DSK2A-B | Phosphorylation of sequences adjacent to AIMs by BIN2 | Promotes the autophagic degradation of BES1 | [197] | |
| DDI1 | No data | |||
| NBR1 | No data |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zientara-Rytter, K.; Subramani, S. The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy. Cells 2019, 8, 40. https://doi.org/10.3390/cells8010040
Zientara-Rytter K, Subramani S. The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy. Cells. 2019; 8(1):40. https://doi.org/10.3390/cells8010040
Chicago/Turabian StyleZientara-Rytter, Katarzyna, and Suresh Subramani. 2019. "The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy" Cells 8, no. 1: 40. https://doi.org/10.3390/cells8010040