1. Introduction
MicroRNA (miRNA) is short noncoding (functional) RNA whose primary function is mRNA degradation and disruption of translation [
1]. Thus, it is generally expected that the primary effect of miRNA transfection (or overexpression) on mRNA expression is suppression. Based on this assumption, numerous miRNA transfection and/or overexpression experiments have been conducted to identify genes that are directly targeted by miRNAs [
2]; during these analyses, only genes with expression levels inversely related to those of miRNA have been sought. Nevertheless, it was found that many mRNAs whose expression was likely to be altered by miRNA transfection and/or overexpression turned out to positively correlate with miRNA expression. For example, Khan et al. [
3] identified multiple genes that are upregulated by miRNA transfection. They reasoned that this effect means competition with endogenous miRNAs because upregulated genes were often targeted by endogenous miRNAs. The protein machinery that binds to endogenous miRNAs was occupied by the transfected miRNAs, and as a result, the genes targeted by endogenous miRNAs were upregulated [
4]. In addition, Carroll et al. [
5] identified a positive correlation between mRNA expression and transfected miRNA. They theorized that the positive correlations are mediated by interactions with transcription factor E2F1.
Despite these findings, to my knowledge, sequence-nonspecific off-target regulation by miRNA transfection has not been extensively studied to date [
2]. Most of the miRNA transfection and/or overexpression experiments have been aimed at identifying canonical targets of miRNAs. Most of these experiments have not been analyzed in the context of sequence-nonspecific off-target regulation by miRNA transfection. Although it is unclear why no one has tried to systematically investigate sequence-nonspecific off-target regulation mediated by miRNA transfection, one possible reason is the lack of a suitable methodology. By definition, miRNA transfection experiments cannot be composed of many samples. Typically, a pair of data points consists of miRNA-transfected cells and mock-transfected cells. Although a few more biological and/or technical replicates are possible, the number of samples available is usually less than 10. This number is often too small to detect significantly altered expression of mRNAs whose total number is up to
. In the case when the aim of a study is identification of canonical interactions between miRNA and mRNA, additional information that can reduce the number of mRNAs under study, e.g., bioinformatically predicted mRNAs targeted by transfected miRNAs, is available. This information can enable researchers to identify significant correlations between transfected miRNAs and mRNAs. Nonetheless, this kind of information is usually not available for the analysis of sequence-nonspecific off-target regulation by miRNA transfection.
In this study, with the aim to resolve this difficulty, I applied tensor decomposition (TD)-based unsupervised feature extraction (FE) to miRNA transfection experiments. TD-based unsupervised FE [
6,
7,
8,
9,
10] is an extension of principal component analysis (PCA)-based unsupervised FE [
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33], which can identify critical genes even when there is only a small number of samples. TD-based unsupervised FE can also identify critical genes by means of a small number of samples available. Because of the use of this methodology, genes whose mRNA expression was likely to be altered by miRNA transfection were identified here in various combinations of cell lines and transfected miRNAs. Most of sets of genes significantly overlapped with one another regardless of transfected cell types and types of miRNA. These genes also showed altered mRNA expression under the influence of a Dicer knockout (KO). This finding suggests that the primary factor that mediated sequence-nonspecific side effects of miRNA transfection is competition for protein machinery with endogenous miRNAs, as suggested by Khan et al. [
3]. In addition, these sets of genes significantly overlapped with various sets of genes whose biological functions and significance have been validated experimentally. Thus, sequence-nonspecific off-target regulation caused by miRNA transfection is expected to also play critical roles in various biological processes.
3. Results
To demonstrate the usefulness of TD-based unsupervised FE, I applied it to artificial data composed of a three-mode tensor,
, which is the expression level of the
ith gene of the
jth sample, where
is control and
is a treated (transfected with distinct miRNAs) sample. Among
N genes,
genes are affected by miRNA transfection while the other
genes are not affected. Among the
genes affected by miRNA transfection,
genes are supposed to be regulated independently of samples (hence, a sequence-nonspecific off-target effect) while the other
genes vary from sample to sample (i.e., miRNA-specific regulation). Applying TD-based unsupervised FE to the artificial dataset (averaged across 100 independent trials), I got a result (
Figure 1).
(
Figure 1A) and
(
Figure 1B), which are always associated with core tensor
with the largest absolute values, represent constant gene expression across
M samples and inverted expression levels between control (
) and treated (
) samples. Accordingly, genes associated with these two are expected to represent sample- or transfected miRNA-independent (thus, sequence-nonspecific off-target) regulation. Given that
is always associated with core tensor
with the largest absolute values,
P-values (
Figure 1C) are computed using
. It is obvious that there is a sharp peak at the smallest
P-values in the histogram of
, which presumably does not correspond to the null hypothesis (that
follows the normal distribution). To test whether genes associated with these much smaller
P-values include genes
, the probabilities to be selected by TD-based unsupervised FE are averaged across
and
, respectively. Then, the former is as large as 0.86, while the latter is as small as 0. (This means that genes
have never been selected by TD-based unsupervised FE.) This observation suggests that TD-based unsupervised FE is effective at sorting out genes—that are expressed independently of samples—from genes expressed only in a limited number of samples and genes not expressed at all. To determine whether TD-based unsupervised FE can outperform the conventional supervised method, the
t test and significance analysis of microarrays (SAM) [
37] were carried out. For these two methods,
P-values obtained were also corrected by means of the BH criterion, and genes associated with adjusted
P-values less than 0.01 were selected. Then, the average probability for
is 0.43 according to the
t test and 0.62 according to SAM. The average probability for
is
according to the
t test and
according to SAM. Therefore, TD-based unsupervised FE is more effective than either the
t test or SAM.
To identify genes whose expression alteration is likely to be mediated by sequence-nonspecific off-target regulation caused by miRNA transfection, integrated analysis of gene expression profiles after transfection of various miRNAs was performed. Simultaneous analysis of multiple experiments each of which employs single miRNA transfection will blur sequence-specific regulation of mRNAs while a sequence-nonspecific off-target effect will remain. Furthermore, to avoid biases due to research groups or individual studies, 11 experiments collected from studies involving distinct combinations of transfected miRNAs and cell lines were analyzed simultaneously (
Table 1).
Genes—whose expression alteration is likely to be caused by sequence-nonspecific off-target regulation that was induced similarly by various transfected miRNAs—were selected using TD-based unsupervised FE in each of the 11 experiments. The reason why TD-based unsupervised FE was employed is as follows. First, PCA-based unsupervised FE, from which TD-based unsupervised FE was developed, is known to function even when only a small number of samples is available [
16,
26]. Tensor representation is also more suitable for the present experiments, where multiple genes, multiple miRNAs and controls, or transfected samples are simultaneously considered (they can be represented as a three-mode tensor; see Methods). Second, we can check whether there are miRNAs associated with a common expression pattern among all the miRNA transfection experiments by studying outcomes; we have opportunities to exclude experiments not associated with sequence-nonspecific off-target regulation caused by miRNA transfection.
These 11 analyzed experiments—in which mRNAs associated with sequence-nonspecific off-target regulation caused by miRNA transfection were successfully identified—deal with distinct cell lines into each of which distinct miRNAs were transfected. Nevertheless, gene sets identified in individual experiments not only significantly overlapped with one another but also were associated with a large enough odds ratio (from 300 to 500,
Table 2), although the number of genes detected in each experiment varied from ∼100 to ∼800 (“#” in
Table 2). This finding suggests that there are some sets of genes whose expression was robustly altered via sequence-nonspecific off-target regulation that was induced similarly by various transfected miRNAs.
Although this finding itself is remarkable enough to be reported, the observed coincidences may be accidental for some unknown reason and may not be associated with anything biologically valid. To validate biological significance of the identified genes, they were uploaded to Enrichr [
38], which is an enrichment analysis server validating various biological terms and concepts. As a result, these genes were found to be enriched with various biological terms and concepts (see below).
First, the identified gene sets mostly included various target genes of transcription factors (TFs) (
Table 3). Although the number of TFs detected varied from ∼10 to ∼100 (#2 in
Table 3), the detection of multiple instances of enrichment with genes that TFs target may be evidence that these genes cooperatively function in the cell because common TFs’ target genes often have shared biological functions [
39,
40]. In particular, the most frequent cases of enrichment of TFs are common among the 11 experiments analyzed. These include EKLF, MYC, NELFA, and E2F1. These data can be biologically interpreted as follows.
The apparent significant alteration of expression of MYC target genes may be due to miRNAs regulating
MYC [
41,
42,
43]. The apparent alteration of expression of E2F1 target genes may be explained similarly because these miRNAs also target
E2F1 [
41,
43]. In actuality, 1551 genes targeted by only one miRNA but associated with altered gene expression caused by miRNA transfection are significantly targeted by MYC and E2F1. Enrichment with EKLF target genes among these 1551 genes not targeted by more than one miRNA—but showing altered expression caused by transfection with one of miRNAs—is a puzzle, although Yien and Beiker suggested that some miRNAs are likely also regulated by EKLF [
44]. Identification of NELFA target genes is explained by the tight interaction between NELFA and c-MYC [
45,
46].
Additionally, I checked “TargetScan microRNA” in Enrichr to test whether enrichment with genes targeted by the transfected miRNAs would be detected. As a result, only for two of the 11 experiments (Experiments No. 2 and 3), enrichment with genes targeted by nonzero miRNAs was detected. This is possibly because
for these two experiments also detected genes targeted by transfected miRNAs (
Figures S2 and S3). In any case, the fact that most of experiments (9 out of 11) did not show enrichment with genes targeted by transfected miRNAs is consistent with the hypothesis that gene expression alteration caused by miRNA transfection is primarily due to the competition for protein machinery between endogenous miRNAs and transfected miRNAs; this pattern can remain unchanged among transfection experiments with different miRNAs.
Next, I checked KEGG pathway enrichment data. Primary enriched KEGG pathways among the identified genes are related to diseases (
Table 4). Seven ((ii), (iii), (v), (vi), (vii), (viii), and (x)) out of 10 most frequently enriched KEGG pathways in the 11 experiments are directly related to various diseases. Among the remaining three ((i), (iv), and (ix)), oxidative phosphorylation is a disease-related KEGG pathway because its malfunction causes combined oxidative phosphorylation deficiency (
https://www.omim.org/entry/609060). Another one, “endoplasmic reticulum”, is also a disease-related pathway because its malfunction is observed in neurological diseases [
47]. Therefore, these genes may also contribute to the onset or progression of various diseases and could be therapeutic targets. As a result, sequence-nonspecific off-target regulation caused by miRNA transfection may be a therapeutic method.
Actually, gene expression alteration mediated by sequence-nonspecific off-target regulation is analogous to treatments with various candidate drugs (
Table 5 and
Table 6). Thus, combinatorial transfection with miRNAs may replace drug treatment in some conditions and can be used for therapeutic purposes, too. For example, LDN-192189 ((ii) in
Table 5) is reported to improve neuronal conversion of human fibroblasts [
48] and was proposed as a therapy for Alzheimer’s disease [
49]. GSK-1059615b ((i) and (iii) in
Table 5) was once considered a PI3K-kt pathway inhibitor in clinical development for the treatment of cancers [
50]. WYE-125132 ((iv) in
Table 5) is also known to suppress tumor growth [
51] (although the name WYE-125132 does not appear in that article, compound 8a was named WYE-125132 later). Afatinib ((v) in
Table 5) is a famous drug for non-small cell lung cancer [
52]. PI-103 ((vi) in
Table 5) is a drug for acute myeloid leukemia [
53]. PD-0325901 was reported to affect heart development [
54]. Chelerythrine chloride ((viii) in
Table 5) has been reported to affect embryonic chick heart cells [
55]. GDC-0980 ((i) in
Table 6) is a known anticancer drug [
56]. PLX-4720 ((ii) in
Table 6) has been considered for both cancer and heart disease treatment [
57]. Dinaciclib is another anticancer drug [
58]. Because these are related to diseases reported in
Table 4, sequence-nonspecific off-target regulation caused by miRNA transfection may be a therapeutic strategy.
Next, I studied tissue specificity of the gene expression alteration caused by sequence-nonspecific off-target regulation that was induced by miRNA transfection. Associations with GTEx tissue samples were also observed. For example, in GTEx up (
Table 7), enrichment cases were observed in the brain and testes, both of which were reported to be outliers in clustering analysis [
59]. Thus, it is not surprising that cases of enrichment in these two tissues were identified primarily. On the other hand, in GTEx down (
Table 8), enrichment instances in the skin were mostly observed. Readers may wonder how sequence-nonspecific off-target regulation caused by miRNA transfection can contribute to the tissue specificity of gene expression profiles. Võsa et al. [
60] observed that polymorphisms in miRNA response elements (MRE-SNPs) that either disrupt a miRNA-binding site or create a new miRNA-binding site can affect the allele-specific expression of target genes. Therefore, sequence-nonspecific off-target regulation caused by miRNA transfection may have the ability to contribute to tissue specificity of the gene expression profiles through distinct functionality of genetic variations in distinct tissues. Despite these coincidences, how the sequence-nonspecific off-target effect contributes to differentiation is unclear. These coincidences may simply be consequences, not causes. More studies are needed to directly implicate the sequence-nonspecific off-target effect in differentiation itself.
Genes whose expression alteration is likely to be caused by sequence-nonspecific off-target regulation that miRNA transfection induces are also enriched in pluripotency (
Table 9). Because miRNAs are known to mediate reprogramming [
61], sequence-nonspecific off-target regulation caused by transfection of multiple miRNAs may contribute to reprogramming too. In actuality, primary binding TFs include MYC ((i) and (iv) in
Table 9) and KLF4 ((vi) and (vii) in
Table 9), which are two of four Yamanaka factors that mediate pluripotency. Other TFs in
Table 9 are DMAP1 (iii) and TIP60 (v). DMAP1 is a member of the TIP60-p400 complex that maintains embryonic stem cell pluripotency [
62]. The remaining one, ZFX (x), has been reported to control the self-renewal of embryonic and hematopoietic stem cells [
63]. In addition, two gene KO experiments have been conducted ((viii) and (ix)). One of the genes in question,
SUZ12, encodes a polycomb group protein that mediates differentiation [
64], whereas the other,
ZFP281, is a known pluripotency suppressor [
65]. There are no known widely accepted mechanisms by which miRNA transfection can induce pluripotency. Because sequence-nonspecific off-target regulation seems to alter expression of genes critical for pluripotency, it may contribute to the mechanism.
Besides, I checked whether protein–protein interactions (PPIs) are enriched in each of the 11 gene sets selected for each of the 11 experiments (
Table 10). Gene sets were uploaded to the STRING server [
66]. For all the gene sets, instances of PPI enrichment were highly significant. Given that proteins rarely function alone and often function in groups, this is evidence that our analysis is biologically reliable.
I demonstrated enrichment of various biological processes in gene sets. Although there were more cases of enrichment of biological terms in Enrichr, it is impossible to consider and discuss all of them. Instead, I discuss the enrichment cases identified in GeneSigDBs in Enrichr that include various biological properties (
Table 11). Numerous GeneSigDBs that reflect a wide range of functional genes were enriched too. This result also suggested that the identified genes may contribute to a wide range of biological activities.
As readers can see, top four most frequently significant terms are related to either diseases or differentiation, which are often said to be biological concepts to which miRNAs contribute to. Although canonical target genes of miRNA have mainly been sought to understand biological functions of miRNAs, sequence-nonspecific off-target regulation may be important as well.
Figure 2 shows the summary of results obtained in this section.
4. Discussion
Many biological conceptual cases of enrichment were observed in the identified sets of genes across the 11 experiments. Nonetheless, detailed mechanisms by which sequence-nonspecific off-target effects (that transfected miRNAs produce) can regulate expression of these genes are unclear. To identify such a mechanism, enrichment of genes associated with the altered expression pattern caused by a Dicer KO within these 11 gene sets was studied by means of Enrichr. Among 16 experiments included in Enrichr (“Single Gene Perturbations from GEO up” and “Single Gene Perturbations from GEO down”), most of them are associated with enrichment of the identified genes in 11 miRNA transfection experiments (
Table 12). In addition, these sets of genes significantly overlap with the set of genes associated with binding to Dicer according to immunoprecipitation (IP) experiments (
Table 12). There are also data supporting the hypothesis that sequence-nonspecific off-target regulation is due to the competition for protein machinery between transfected miRNAs and endogenous miRNAs in the cell.
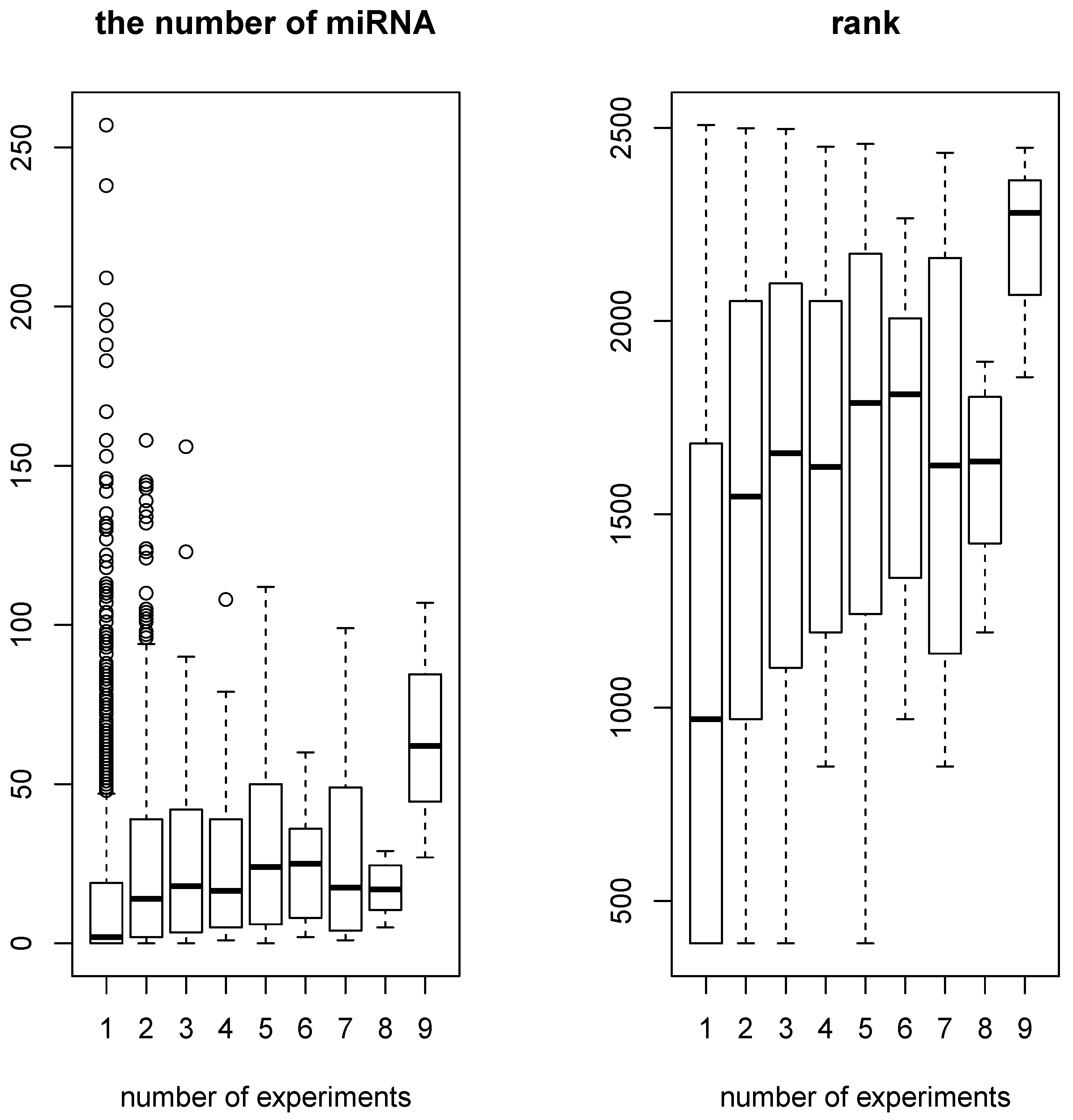
On the other hand, the number of miRNAs that target genes whose expression was likely to be altered by miRNA transfection significantly correlated with the number of experiments where individual genes were selected among the 11 conducted experiments (see
Figure 3. Pearson’s and Spearman’s correlation coefficients are 0.13,
and 0.29,
, respectively). Genes targeted by a greater number of individual miRNAs are likely to be affected by miRNA transfection, which occupies the protein machinery binding to transcripts of these genes. Therefore, the significant correlation is consistent with the hypothesis that sequence-nonspecific off-target regulation is due to competition for protein machinery between a transfected miRNA and endogenous miRNAs in cells.
Thus, primarily, gene expression alteration by sequence-nonspecific off-target regulation caused by miRNA transfection is likely due to suppression of miRNA functionality because of a reduction in the amount of available protein machinery owing to occupation by transfected miRNAs.
Despite the above arguments, it cannot be proven that competition for protein machinery is the primary cause of sequence-nonspecific off-target regulation. Upregulation of genes can also be caused by indirect effects, e.g., genes that suppress expression of other genes are repressed by transfected miRNA, although this mechanism is unlikely to cause upregulation of common genes regardless of the transfected miRNAs. Additional experimental validation will clarify which one is the correct scenario.
Furthermore, I compared the performance of TD-based unsupervised FE with the performance of major supervised methods. Previously, when PCA-based unsupervised FE, on which TD-based unsupervised FE is based, has been applied to various problems, PCA-based unsupervised FE often outperformed other (supervised) methods. For example, when PCA-based unsupervised FE was successfully used to identify genes commonly associated with aberrant promoter methylation among three autoimmune diseases [
19], no supervised methods—except for PCA-based unsupervised FE—could identify common genes. On the other hand, when PCA-based unsupervised FE was applied to identify common HDACi target genes between two independent HDACis [
16], two supervised methods (Limma [
67] and categorical regression, i.e. analysis of variance (ANOVA)) identified the same number of common genes as did PCA-based unsupervised FE. Nevertheless, biological validation of the selected genes supported the superiority of PCA-based unsupervised FE. In contrast to the many instances of biological-term enrichment that were observed among genes selected by PCA-based unsupervised FE, such cases of enrichment among the genes selected by a supervised method were not detected.
Although these are only two examples, this kind of advantages of PCA-based unsupervised FE have often been observed. To confirm the superiority of TD-based unsupervised FE too, I consider Experiments No. 9 and No. 10 in
Table 1 for the comparisons with other (supervised) methods. The reason for this choice is as follows. At first, these two were taken from the same dataset (GSE37729) and the same miRNAs (miR-107/181b) were transfected; thus, these two experiments are expected to have a greater number of common genes selected than do other pairs of experiments in
Table 1. Second, the pair No. 9 and No. 10 has the highest odds ratio in
Table 2, as expected. Thus, these data are suitable for the comparison with another method.
When using ANOVA and SAM [
37], I found that
P-values and the odds ratio computed by Fisher’s exact test are
and
, respectively, for both methods (the number of genes selected is taken to be 103 and 104 for Experiments No. 9 and No. 10, respectively, using ranking based upon
P-values assigned to each gene because these numbers are the same as those selected by TD-based unsupervised FE, see “#” in
Table 2).
A more sophisticated and advanced supervised method may show somewhat better performance than do SAM and categorical regression. Because TD-based unsupervised FE highly outperformed these two conventional and frequently used supervised methods, it is unlikely that another supervised method can compete with TD-based unsupervised FE (advantages of PCA-based unsupervised FE over various conventional supervised methods have been reported repeatedly too [
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33]). More comprehensive performance comparisons with the
t test are provided in the
Supplementary Materials.
Readers may also wonder whether the null assumption that
obeys a normal distribution is appropriate because
is not proven to follow a normal distribution. This approach is not problematic for the following reasons. First, the null hypothesis that
obeys a normal distribution is supposed to be rejected later. Thus, even if
does not follow the normal distribution, this is not a problem. Second, it is reasonable to assume that
follows the normal distribution under the assumption that
is drawn from a random number (
Figure 1C); this statement is suitable as the null hypothesis. Be that as it may, a question may arise whether the null hypothesis that
obeys the normal distribution is not suitable if this assumption is mostly violated. To evaluate how well the null hypothesis is fulfilled, I demonstrate the result for GSE26996 as a typical example.
Figure 4A presents the scatter plot of
where
served for selection of genes as mentioned in the
Section 2.5.1. Considering the fact that the total number of probes in the microarray is more than 20,000 and the number of probes selected is 379, these 379 probes are obviously outliers along the direction of
.
Figure 4B depicts the histogram of
under the null hypothesis that
follows the normal distribution. Although smaller
(<0.3) s deviate from constant values that are expected under the null hypothesis, a sharp peak that includes the selected 379 probes is evidently located at the largest
. Consequently, it is satisfactory for identifying genes
i associated with much larger (that is, invalidating the null hypothesis) absolute
values.
Finally, I would like to consider possible objections to the hypothesis proposed in this study: sequence-nonspecific off-target regulation of mRNA mediated by miRNA transfection is primarily mediated by competition for the protein machinery. The first possible objection is that some miRNA can bind to a promoter region directly. This process in not mentioned in the above discussion. For example, Kim et al. [
68] found that miR-320 can bind to the promoter region of
POLR3D. Nevertheless, this kind of direct binding to DNA by transfected miRNA cannot be the interpretation of the present findings, because it is still sequence specific. Thus, direct binding to DNA cannot be an alternative interpretation of the sequence-nonspecific regulation presented in
Table 2. The second objection is that miRNA can sometimes bind to mRNA with insufficient support by proteins. For example, Lima et al. [
69] found that single-stranded siRNAs can bind to mRNA. Given that single-stranded siRNAs do not have to be processed by the DICER that is mentioned in
Table 12, this topic apparently seems to be outside the scope of this study. Nonetheless, single-stranded siRNAs that Lima et al. identified still need the AGO2 protein. Thus, the regulation of mRNA expression by single-stranded siRNAs can still be under the control of competition for protein machinery. Therefore, it is hard to say whether the process identified by Lima et al. is independent of protein machinery competition. The third objection to the scenario proposed in this study is that miRNA can often upregulate target mRNAs [
70,
71]. This means that observation of upregulation caused by miRNA transfection—which was brought up as one of side proofs for protein machinery competition in the text above—does not always have to be mediated by competition for protein machinery. On the other hand, this process is also sequence specific. Thus, direct upregulation by transfected miRNA still cannot explain the sequence-nonspecific regulation of mRNAs presented in
Table 2. Therefore, at the moment, protein machinery competition is the only possible explanation of the sequence-nonspecific regulation of mRNAs shown in
Table 2.