Elucidating the Role of MicroRNA-18a in Propelling a Hybrid Epithelial–Mesenchymal Phenotype and Driving Malignant Progression in ER-Negative Breast Cancer
Abstract
:1. Background
2. Materials and Methods
2.1. Cell Lines, Culture and Transfection with miR-18a Synthetic Inhibitors
2.2. Protein Expression Analysis by Western Blot
2.3. Immunophenotyping by Flow-Cytometry
2.4. Dual Immunofluorescence
2.5. Computational Analysis for Correlation with E/M Hybrid Score
2.6. Breast Tumour Specimens Used for Gene Expression Analysis
2.7. mRNA and miRNA Expression Analysis Using Quantitative PCR
2.8. Analysis of Mutational Spectrum of Breast Tumours of the METABRIC Cohort
2.9. Analysis of Differentially Expressed Genes (DEGs) and Pathways in Breast Tumours of the TCGA and METABRIC Series
2.10. Correlative Analysis of Published EMT Scores with miR-18a Expression in Breast Tumours of the TCGA and METABRIC Series
2.11. In Vitro Cell Migration—Wound Closure Assay
2.12. Immunohistochemistry of Residual Tumours to Evaluate Expression of Integrin β3
2.13. Evaluation of Drug Cytotoxicity Using MTT
2.14. Generation of Mammospheres and Extreme Limiting Dilution Assay (ELDA) to Assess Clonogenicity
2.15. Estimate Analysis and Immune Cell Identification
2.16. Breast Xenograft in In Vivo Studies
2.17. Histopathological Analysis and Immunostaining of Mice-Derived Tumours
2.18. miR-18a Target Prediction
2.19. Statistical Analysis
3. Results
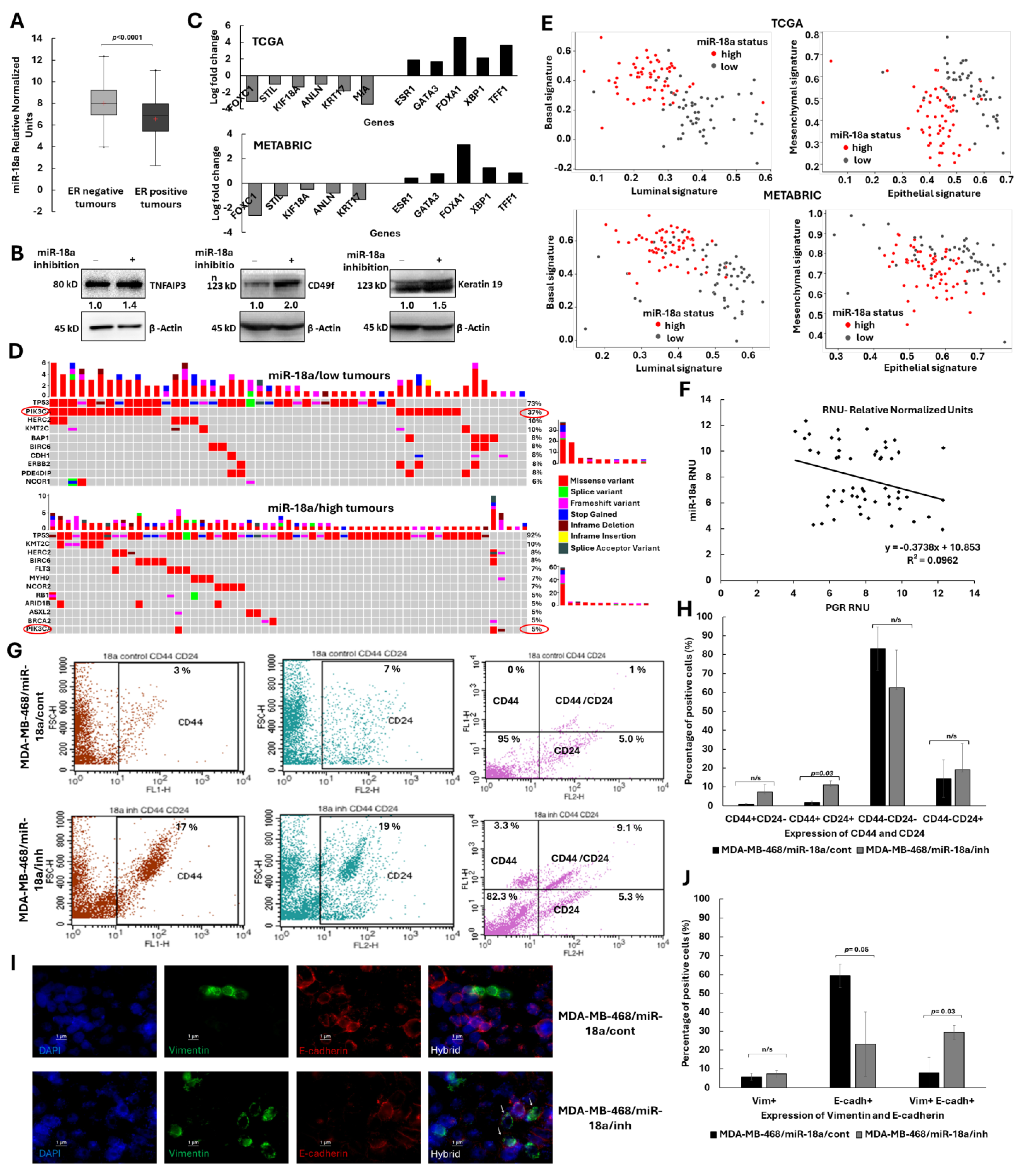
3.1. Low Levels of miR-18a Enriches for the Hybrid Epithelial/Mesenchymal–Lumino/Basal Phenotype in ER-Negative Breast Cancer
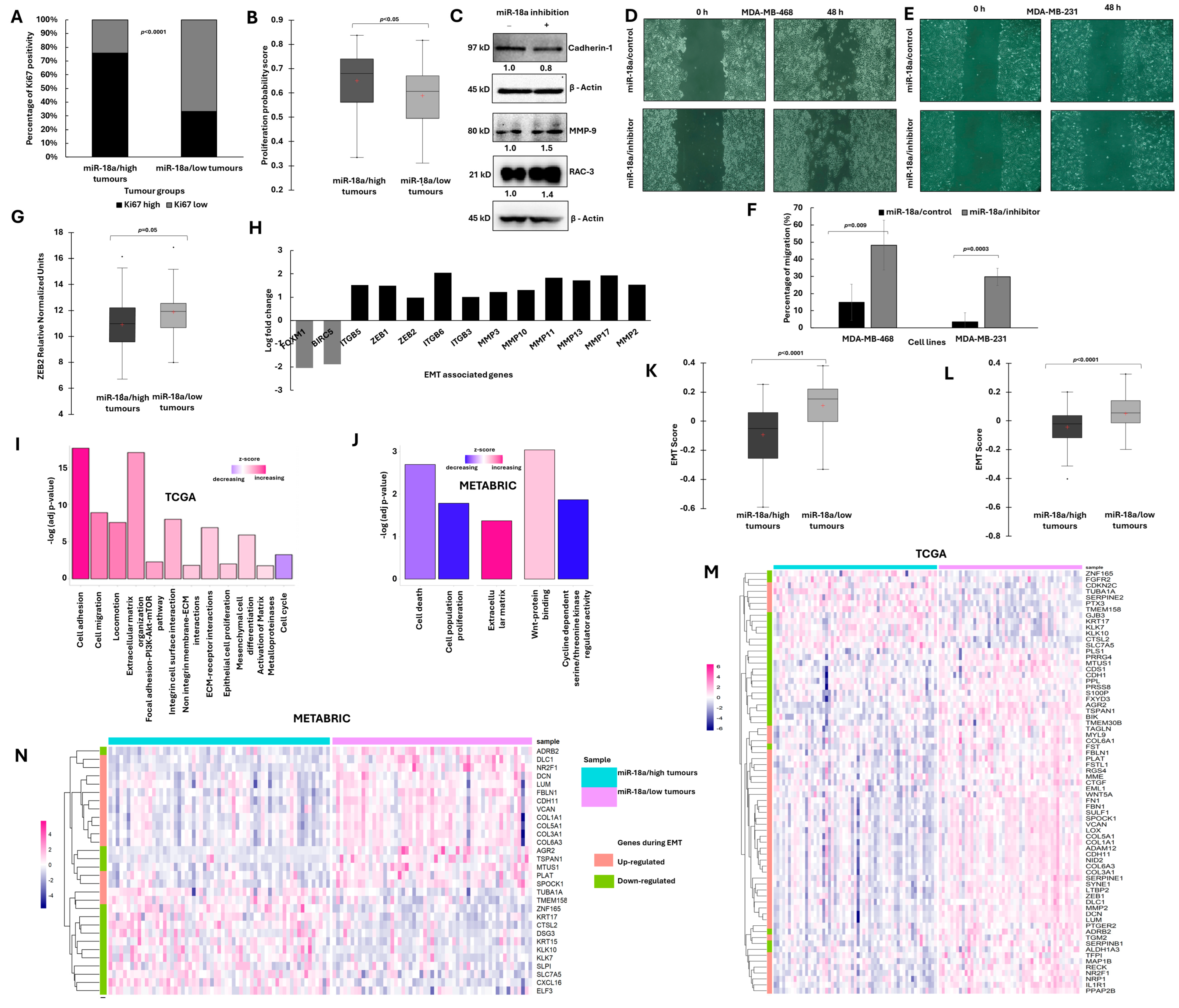
3.2. ER-Negative Breast Cancer with Low miR-18a Is Associated with Low Proliferation and Enhanced EMT Traits
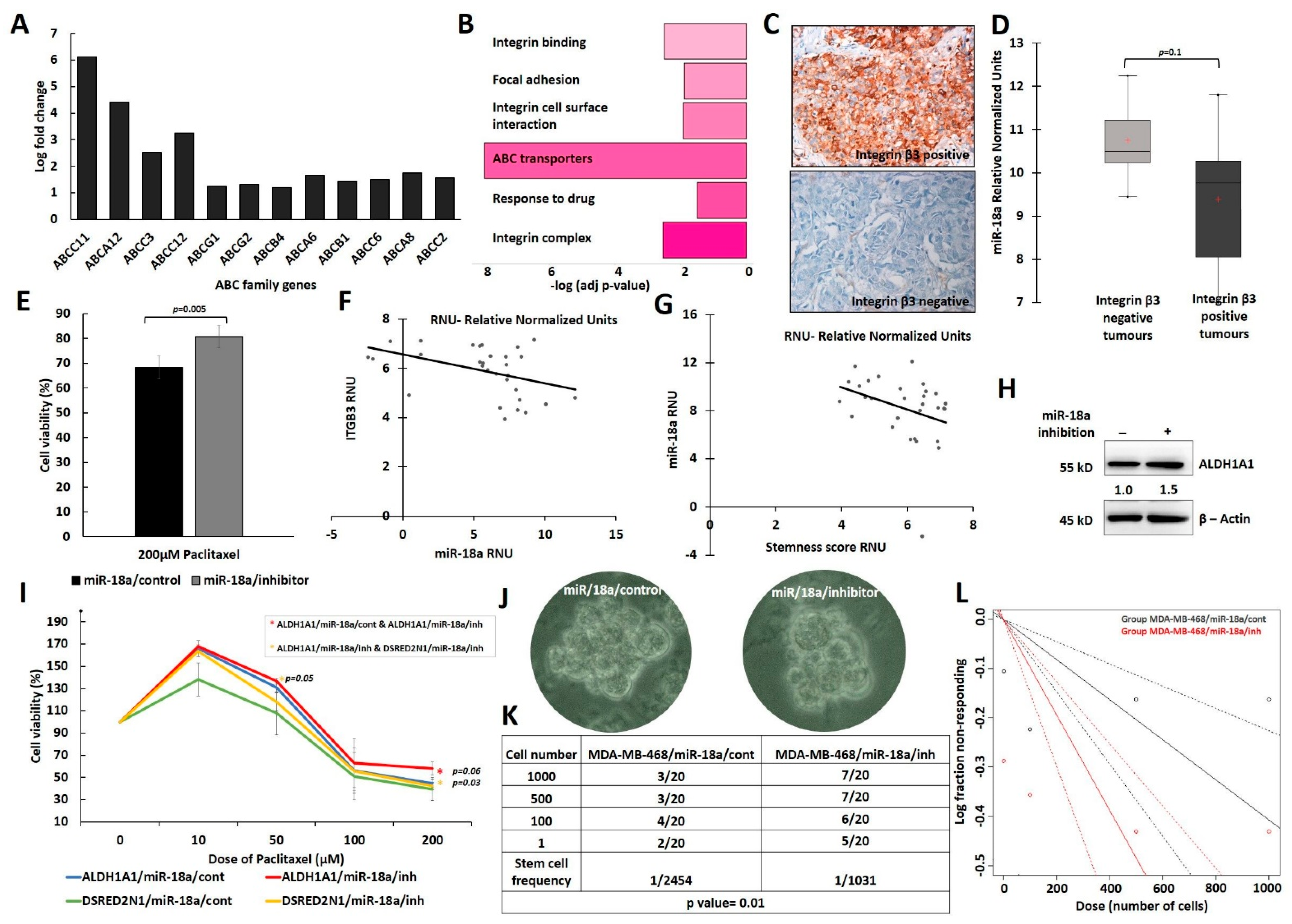
3.3. Low Levels of miR-18a Are Associated with Increased Chemoresistance and Cancer Stemness in the ER-Negative Subtype
3.4. Lower Levels of miR-18a in ER-Negative Breast Cancer Correlates with Increased Stromal-Immune Infiltration and Immunosuppression
3.5. HIF-1α Inhibition Leads to a Reversal of Hybrid E/M Phenotype in miR-18a Inhibited Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| E/M | Epithelial/Mesenchymal |
| miRNAs | microRNAs |
| ELDA | Extreme limiting dilution assay |
| EMT | Epithelial to Mesenchymal Transition |
| E/M hybrid | Epithelial–Mesenchymal Hybrid |
| EMP | Epithelial–Mesenchymal Plasticity |
| NCCS | National Centre for Cell Science |
| ATCC | American Type Culture Collection |
| PFA | Paraformaldehyde |
| FFPE | Formalin-fixed paraffin-embedded |
| QC | Quality control |
| NACT | Neoadjuvant chemotherapy |
| IHC | Immunohistochemistry |
| DEGs | Differentially Expressed Genes |
| FGF | Fibroblast growth factor |
| EGF | Epidermal growth factor |
| CSC | Cancer stem cell |
| ESTIMATE | Estimation of Stromal and Immune cells in Malignant Tumour tissues using Expression data |
References
- Kuukasjärvi, T.; Kononen, J.; Helin, H.; Holli, K.; Isola, J. Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J. Clin. Oncol. 1996, 14, 2584–2589. [Google Scholar] [CrossRef]
- Zattarin, E.; Leporati, R.; Ligorio, F.; Lobefaro, R.; Vingiani, A.; Pruneri, G.; Vernieri, C. Hormone Receptor Loss in Breast Cancer: Molecular Mechanisms, Clinical Settings, and Therapeutic Implications. Cells 2020, 9, 2644. [Google Scholar] [CrossRef]
- Putti, T.C.; El-Rehim, D.M.A.; Rakha, E.A.; Paish, C.E.; Lee, A.H.S.; Pinder, S.E.; Ellis, I.O. Estrogen receptor-negative breast carcinomas: A review of morphology and immunophenotypical analysis. Mod. Pathol. 2005, 18, 26–35. [Google Scholar] [CrossRef]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [CrossRef]
- Lindström, L.S.; Yau, C.; Czene, K.; Thompson, C.K.; Hoadley, K.A.; Van’t Veer, L.J.; Balassanian, R.; Bishop, J.W.; Carpenter, P.M.; Chen, Y.Y.; et al. Intratumor Heterogeneity of the Estrogen Receptor and the Long-term Risk of Fatal Breast Cancer. J. Natl. Cancer Inst. 2018, 110, 726–733. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4. [Google Scholar] [CrossRef]
- Hou, J.; Ye, X.; Wang, Y.; Li, C. Stratification of Estrogen Receptor-Negative Breast Cancer Patients by Integrating the Somatic Mutations and Transcriptomic Data. Front. Genet. 2021, 12, 610087. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Hong, H.-C.; Chuang, C.-H.; Huang, W.-C.; Weng, S.-L.; Chen, C.-H.; Chang, K.-H.; Liao, K.-W.; Huang, H.-D. A panel of eight microRNAs is a good predictive parameter for triple-negative breast cancer relapse. Theranostics 2020, 10, 8771–8789. [Google Scholar] [CrossRef]
- Qattan, A. Novel miRNA Targets and Therapies in the Triple-Negative Breast Cancer Microenvironment: An Emerging Hope for a Challenging Disease. Int. J. Mol. Sci. 2020, 21, 8905. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012, 31, 653–662. [Google Scholar] [CrossRef]
- Ye, X.; Brabletz, T.; Kang, Y.; Longmore, G.D.; Nieto, M.A.; Stanger, B.Z.; Yang, J.; Weinberg, R.A. Upholding a role for EMT in breast cancer metastasis. Nature 2017, 547, E1–E3. [Google Scholar] [CrossRef]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Lamperska, K. Good or not good: Role of miR-18a in cancer biology. Rep. Pr. Oncol. Radiother. 2020, 25, 808–819. [Google Scholar] [CrossRef]
- Shen, K.; Cao, Z.; Zhu, R.; You, L.; Zhang, T. The dual functional role of MicroRNA-18a (miR-18a) in cancer development. Clin. Transl. Med. 2019, 8, 32. [Google Scholar] [CrossRef]
- Nair, M.G.; Prabhu, J.S.; Korlimarla, A.; Rajarajan, S.; Hari, P.S.; Kaul, R.; Alexander, A.; Raghavan, R.; Srinath, B.S.; Sridhar, T.S. miR-18a activates Wnt pathway in ER-positive breast cancer and is associated with poor prognosis. Cancer Med. 2020, 9, 5587–5597. [Google Scholar] [CrossRef]
- Nair, M.G.; Snijesh, V.P.; Patil, S.; Anupama, C.E.; Mukherjee, G.; Kumar, R.V.; Prabhu, J.S.; Sridhar, T.S. miR-18a Mediates Immune Evasion in ER-Positive Breast Cancer through Wnt Signaling. Cells 2022, 11, 1672. [Google Scholar] [CrossRef]
- Krutilina, R.; Sun, W.; Sethuraman, A.; Brown, M.; Seagroves, T.N.; Pfeffer, L.M.; Ignatova, T.; Fan, M. MicroRNA-18a inhibits hypoxia-inducible factor 1α activity and lung metastasis in basal breast cancers. Breast Cancer Res. 2014, 16, R78. [Google Scholar] [CrossRef]
- Sha, L.Y.; Zhang, Y.; Wang, W.; Sui, X.; Liu, S.K.; Wang, T.; Zhang, H. MiR-18a upregulation decreases Dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2201–2208. [Google Scholar]
- Nair, M.G.; Desai, K.; Prabhu, J.S.; Hari, P.S.; Remacle, J.; Sridhar, T.S. β3 integrin promotes chemoresistance to epirubicin in MDA-MB-231 through repression of the pro-apoptotic protein, BAD. Exp. Cell Res. 2016, 346, 137–145. [Google Scholar] [CrossRef]
- Freeberg, M.A.; Fromont, L.A.; D’Altri, T.; Romero, A.F.; Ciges, J.I.; Jene, A.; Kerry, G.; Moldes, M.; Ariosa, R.; Bahena, S.; et al. The European Genome-phenome Archive in 2021. Nucleic Acids Res 2022, 50, D980–D987. [Google Scholar] [CrossRef] [PubMed]
- Aftimos, P.; Azim, H.A.; Sotiriou, C. Chapter 26—Molecular Biology of Breast Cancer. In Molecular Pathology, 2ed Edition; Coleman, W.B., Tsongalis, G.J., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 569–588. [Google Scholar]
- Jia, R.; Li, Z.; Liang, W.; Ji, Y.; Weng, Y.; Liang, Y.; Ning, P. Identification of key genes unique to the luminal a and basal-like breast cancer subtypes via bioinformatic analysis. World J. Surg. Oncol. 2020, 18, 268. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, J.S.; Korlimarla, A.; Anupama, C.E.; Alexander, A.; Raghavan, R.; Kaul, R.; Desai, K.; Rajarajan, S.; Manjunath, S.; Correa, M.; et al. Dissecting the Biological Heterogeneity within Hormone Receptor Positive HER2 Negative Breast Cancer by Gene Expression Markers Identifies Indolent Tumors within Late Stage Disease. Transl. Oncol. 2017, 10, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, J.S.; Korlimarla, A.; Desai, K.; Alexander, A.; Raghavan, R.; Anupama, C.; Dendukuri, N.; Manjunath, S.; Correa, M.; Raman, N.; et al. A Majority of Low (1–10%) ER Positive Breast Cancers Behave Like Hormone Receptor Negative Tumors. J. Cancer 2014, 5, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Mak, M.P.; Tong, P.; Diao, L.; Cardnell, R.J.; Gibbons, D.L.; William, W.N.; Skoulidis, F.; Parra, E.R.; Rodriguez-Canales, J.; Wistuba, I.I.; et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2016, 22, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Gröger, C.J.; Grubinger, M.; Waldhör, T.; Vierlinger, K.; Mikulits, W. Meta-Analysis of Gene Expression Signatures Defining the Epithelial to Mesenchymal Transition during Cancer Progression. PLoS ONE 2012, 7, e51136. [Google Scholar] [CrossRef]
- Shanmugam, G.; Mohan, A.; Kumari, K.; Louis, J.M.; Soumya Krishnan, U.; Balagopal, P.G.; George, N.A.; Sebastian, P.; Maliekal, T.T. A novel reporter construct for screening small molecule inhibitors that specifically target self-renewing cancer cells. Exp. Cell Res. 2019, 383, 111551. [Google Scholar] [CrossRef]
- Filippi, S.; Paccosi, E.; Balzerano, A.; Ferretti, M.; Poli, G.; Taborri, J.; Brancorsini, S.; Proietti-De-Santis, L. CSA Antisense Targeting Enhances Anticancer Drug Sensitivity in Breast Cancer Cells, including the Triple-Negative Subtype. Cancers 2022, 14, 1687. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.-R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.-Y.; Wang, H.; Guo, A.-Y. ImmuCellAI: A unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv. Sci. 2019, 7, 1902880. [Google Scholar] [CrossRef]
- Xu, L.; Yu, W.; Xiao, H.; Lin, K. BIRC5 is a prognostic biomarker associated with tumor immune cell infiltration. Sci. Rep. 2021, 11, 390. [Google Scholar] [CrossRef]
- Ziegler, Y.; Laws, M.J.; Sanabria Guillen, V.; Kim, S.H.; Dey, P.; Smith, B.P.; Gong, P.; Bindman, N.; Zhao, Y.; Carlson, K.; et al. Suppression of FOXM1 activities and breast cancer growth in vitro and in vivo by a new class of compounds. NPJ Breast Cancer 2019, 5, 45. [Google Scholar] [CrossRef]
- Choi, C.-H. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int. 2005, 5, 30. [Google Scholar] [CrossRef]
- Duvivier, L.; Gerard, L.; Diaz, A.; Gillet, J.-P. Linking ABC transporters to the hallmarks of cancer. Trends Cancer 2024, 10, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Quan, Q.; Wang, X.; Lu, C.; Ma, W.; Wang, Y.; Xia, G.; Wang, C.; Yang, G. Cancer stem-like cells with hybrid epithelial/mesenchymal phenotype leading the collective invasion. Cancer Sci. 2020, 111, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Vaillant, F.; Asselin-Labat, M.L.; Shackleton, M.; Forrest, N.C.; Lindeman, G.J.; Visvader, J.E. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008, 68, 7711–7717. [Google Scholar] [CrossRef] [PubMed]
- Pommier, R.M.; Sanlaville, A.; Tonon, L.; Kielbassa, J.; Thomas, E.; Ferrari, A.; Sertier, A.-S.; Hollande, F.; Martinez, P.; Tissier, A.; et al. Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution. Nat. Commun. 2020, 11, 3431. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Willard-Gallo, K. Tumor-infiltrating follicular helper T cells: The new kids on the block. Oncoimmunology 2013, 2, e26066. [Google Scholar] [CrossRef]
- Yang, L.; Roberts, D.; Takhar, M.; Erho, N.; Bibby, B.A.S.; Thiruthaneeswaran, N.; Bhandari, V.; Cheng, W.C.; Haider, S.; McCorry, A.M.B.; et al. Development and Validation of a 28-gene Hypoxia-related Prognostic Signature for Localized Prostate Cancer. EBioMedicine 2018, 31, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Ye, I.C.; Fertig, E.J.; DiGiacomo, J.W.; Considine, M.; Godet, I.; Gilkes, D.M. Molecular Portrait of Hypoxia in Breast Cancer: A Prognostic Signature and Novel HIF-Regulated Genes. Mol. Cancer Res. 2018, 16, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Qiao, L.; Liao, J.; Liu, Q.; Liu, P.; Liu, L. A novel hypoxia gene signature indicates prognosis and immune microenvironments characters in patients with hepatocellular carcinoma. J. Cell Mol. Med. 2021, 25, 3772–3784. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Russo, J. ERalpha-negative and triple negative breast cancer: Molecular features and potential therapeutic approaches. Biochim. Biophys. Acta 2009, 1796, 162–175. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Wu, C.I.; Shen, Y.; Tang, T. Evolution under canalization and the dual roles of microRNAs: A hypothesis. Genome Res. 2009, 19, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Sahoo, S.; Donnenberg, V.S.; Chakraborty, P.; Jolly, M.K.; Sant, S. P4HA2: A link between tumor-intrinsic hypoxia, partial EMT and collective migration. Adv. Cancer Biol. Metastasis 2022, 5, 100057. [Google Scholar] [CrossRef] [PubMed]
- Kröger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Yeh, S.H.; Lu, C.C.; Yu, S.L.; Chen, H.Y.; Lin, C.Y.; Chen, D.S.; Chen, P.J. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 2009, 136, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarian, H.; Yıldız, M.T.; Tutar, Y. MicroRNA Targeting. Methods Mol. Biol. 2022, 2257, 105–130. [Google Scholar] [CrossRef] [PubMed]
- Baran, A.; Fırat Baran, M.; Keskin, C.; Hatipoğlu, A.; Yavuz, Ö.; İrtegün Kandemir, S.; Adican, M.T.; Khalilov, R.; Mammadova, A.; Ahmadian, E.; et al. Investigation of Antimicrobial and Cytotoxic Properties and Specification of Silver Nanoparticles (AgNPs) Derived From Cicer arietinum L. Green Leaf Extract. Front. Bioeng. Biotechnol. 2022, 10, 855136. [Google Scholar] [CrossRef] [PubMed]
- Ramazanli, V.; Ahmadov, I. Synthesis of Silver Nanoparticles by Using Extract of Olive Leaves. Adv. Biol. Earth Sci. 2022, 7, 238–244. [Google Scholar]
- Bertucci, A.; Kim, K.-H.; Kang, J.; Zuidema, J.M.; Lee, S.H.; Kwon, E.J.; Kim, D.; Howell, S.B.; Ricci, F.; Ruoslahti, E.; et al. Tumor-Targeting, MicroRNA-Silencing Porous Silicon Nanoparticles for Ovarian Cancer Therapy. ACS Appl. Mater. Interfaces 2019, 11, 23926–23937. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Dando, I.; Strippoli, R.; Kumar, S.; Somoza, A.; Cordani, M.; Tafani, M. Nanomaterials for Autophagy-Related miRNA-34a Delivery in Cancer Treatment. Front. Pharmacol. 2020, 11, 1141. [Google Scholar] [CrossRef]
- Sukumar, U.K.; Bose, R.J.C.; Malhotra, M.; Babikir, H.A.; Afjei, R.; Robinson, E.; Zeng, Y.; Chang, E.; Habte, F.; Sinclair, R.; et al. Intranasal delivery of targeted polyfunctional gold-iron oxide nanoparticles loaded with therapeutic microRNAs for combined theranostic multimodality imaging and presensitization of glioblastoma to temozolomide. Biomaterials 2019, 218, 119342. [Google Scholar] [CrossRef]


| Sl No. | Gene | Function (Source—GeneCards, NCBI Gene) |
|---|---|---|
| 1 | HIF1A | Mediates hypoxia-induced expression of mRNA-encoding genes; regulates the expression of non-coding RNAs, which are critical regulators of migration, invasion and metastasis |
| 2 | BBX | Transcription factor necessary for cell cycle progression from G1 to S phase |
| 3 | CDK19 | Mediator kinases, transcriptional co-regulators |
| 4 | DICER1 | Responsible for cleaving double-stranded RNAs into small interfering RNAs and microRNAs |
| 5 | NEDD9 | Positive regulator of epithelial–mesenchymal transition and promotes invasion |
| 6 | EPB41L1 | Role in cell adhesion and migration, malignant progression |
| 7 | ESR1 | Regulates the transcription of estrogen-inducible genes that play a role in growth, metabolism, sexual development, gestation |
| 8 | GLRB | Down-regulation of neuronal excitability, generation of inhibitory postsynaptic currents |
| 9 | INADL | Mediate protein–protein interactions, regulate the formation and stabilization of tight junctions |
| 10 | MAP3K1 | Serine/threonine kinase in multiple cell signalling cascades |
| 11 | PDE4D | Major regulators of cAMP-hydrolyzing activity |
| 12 | PHC3 | Transcriptional repression, chromatin remodelling and modification of histones |
| 13 | RORA | Interacts with NM23-2, a nucleoside diphosphate kinase involved in organogenesis and differentiation, as well as with NM23-1, the product of a tumour metastasis suppressor candidate gene |
| 14 | SH3BP4 | Involved in cargo-specific control of clathrin-mediated endocytosis, specifically controlling the internalization of a specific protein receptor. |
| 15 | ZNF367 | Transcriptionally activates KIF15 and regulates cell cycle |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nair, M.G.; Mavatkar, A.D.; Naidu, C.M.; V. P., S.; C. E., A.; Rajarajan, S.; Sahoo, S.; Mohan, G.; Jaikumar, V.S.; Ramesh, R.S.; et al. Elucidating the Role of MicroRNA-18a in Propelling a Hybrid Epithelial–Mesenchymal Phenotype and Driving Malignant Progression in ER-Negative Breast Cancer. Cells 2024, 13, 821. https://doi.org/10.3390/cells13100821
Nair MG, Mavatkar AD, Naidu CM, V. P. S, C. E. A, Rajarajan S, Sahoo S, Mohan G, Jaikumar VS, Ramesh RS, et al. Elucidating the Role of MicroRNA-18a in Propelling a Hybrid Epithelial–Mesenchymal Phenotype and Driving Malignant Progression in ER-Negative Breast Cancer. Cells. 2024; 13(10):821. https://doi.org/10.3390/cells13100821
Chicago/Turabian StyleNair, Madhumathy G., Apoorva D. Mavatkar, Chandrakala M. Naidu, Snijesh V. P., Anupama C. E., Savitha Rajarajan, Sarthak Sahoo, Gayathri Mohan, Vishnu Sunil Jaikumar, Rakesh S. Ramesh, and et al. 2024. "Elucidating the Role of MicroRNA-18a in Propelling a Hybrid Epithelial–Mesenchymal Phenotype and Driving Malignant Progression in ER-Negative Breast Cancer" Cells 13, no. 10: 821. https://doi.org/10.3390/cells13100821





