Isolation of Myenteric and Submucosal Plexus from Mouse Gastrointestinal Tract and Subsequent Co-Culture with Small Intestinal Organoids
Abstract
:1. Introduction
Experimental Design of a Novel Co-Culture Technique
2. Materials and Methods
2.1. Reagents and Material Setup
2.1.1. Neuron Isolation Reagents
- 1.
- Measure out the appropriate amounts of each compound based on the desired final volume.
- Sodium chloride (NaCl): Causes serious eye irritation. Avoid contact with eyes.
- Magnesium sulfate (MgSO4): Sodium dihydrogen phosphate (NaH2PO4). The substance has warnings for being harmful through ingestion, skin contact, and inhalation due to acute toxicity.
- Calcium chloride (CaCl2): Causes serious eye irritation. Avoid contact with eyes.
- For all: Do not breathe dust. Do not get in eyes, on skin, or on clothing. Wear protective gloves/protective clothing/eye protection/face protection. Wash hands thoroughly after handling.
- 2.
- Dissolve the measured amounts of each compound in double-distilled water (ddH2O). Dissolve them completely to ensure a homogeneous solution.
- 3.
- Adjust the pH of the solution to 7.4 using a pH meter and a suitable pH-adjusting agent such as hydrochloric acid (HCl) or sodium hydroxide (NaOH).
- Hydrochloric acid (HCl): The substance is dangerous, causing severe skin burns, eye damage, and acute toxicity when inhaled.
- Sodium hydroxide (NaOH): Causes severe skin burns and eye damage.
- 4.
- Once the pH is adjusted, check the osmolality of the solution to ensure it is within the desired range. The osmolality should typically be around 280–320 mOsm/kg.
- 5.
- Filter the solution using a sterile filter (pore size of 0.22 μm) to remove any particulate matter or contaminants.
- 6.
- Supplement with the correspondent amount of antibiotic/antimycotic solution.
- 7.
- Transfer the filtered Krebs solution to a sterilized container or aliquot into smaller volumes for convenient use.
- 1.
- Measure out the appropriate amounts of each compound based on the desired final volume.
- 2.
- Dissolve the measured amounts in carbogen-bubbled Krebs solution.
- 3.
- Filter the solution using a sterile filter (pore size of 0.22 μm) to remove any particulate matter or contaminants.
- 4.
- Place on ice until use.
- Antibiotic cocktail (20X) (Table 3):
- Ampicillin sodium: The substance may trigger allergic skin reactions and can also cause allergy or asthma symptoms, leading to breathing difficulties if inhaled.
- Gentamycin: The substance presents various hazards, including the potential for allergic skin reactions, damage to fertility or the unborn child, harm to organs through prolonged exposure, and being very toxic to aquatic life, both acutely and with long-lasting effects.
- Gibco™ Antibiotic/Antimycotic, 100X: Reproductive Harm
- Neomycin: The substance is entirely harmful if swallowed, and it has a full likelihood of causing allergic skin reactions as well as allergy or asthma symptoms when inhaled.
- Metronidazole: The substance carries various hazards, including being harmful if swallowed, in contact with skin, or inhaled, with warnings about acute toxicity. It is also associated with potential genetic defects, carcinogenicity (both suspected and confirmed), and causing damage to organs through prolonged exposure, as well as being hazardous to aquatic life with long-lasting effects.
- Vancomycin hydrochloride: The substance is capable of causing skin irritation, allergic skin reactions, serious eye irritation, and respiratory irritation, prompting warnings for skin, eye, and respiratory effects.
- 1.
- Ampicillin solution: Weigh 1 g of sodium ampicillin and dissolve in 10 mL of ddH2O to achieve a concentration of 100 mg/mL. Filter the ampicillin solution using a sterile filter with a pore size of 0.22 μm by first prewashing the filter with some ddH2O run through it. Aliquot ampicillin solution into 2 mL aliquots and store at −20 °C for a period of one year.
- 2.
- Gibco™ Antibiotic/Antimycotic solution, 100X: Prepare 5 mL aliquots and store them at −20 °C. Gibco™ Antibiotic-Antimycotic solution contains 10,000 units/mL of penicillin, 10,000 µg/mL of streptomycin, and 25 µg/mL of Gibco Amphotericin B to avoid bacterial and fungal contamination.
- 3.
- Metronidazole solution, 50 mg/mL: Weigh 500 mg of metronidazole and dissolve in 10 mL of ddH2O to achieve a concentration of 50 mg/mL. Filter the metronidazole solution using a sterile filter with a pore size of 0.22 μm by first prewashing the filter with some ddH2O run through it. Aliquot metronidazole solution into 2 mL aliquots and store at 4 °C for up to two years.
- 4.
- Vancomycin hydrochloride solution, 200 mM: First, calculate the mass of vancomycin hydrochloride powder required to prepare the desired concentration: Mass (g) = (Concentration (M) × Volume (L) × Molecular weight (g/mol)). In this case, the concentration is 200 mM (0.2 M), the volume is 500 μL (0.0005 L), and the molecular weight of vancomycin hydrochloride is 1485.71 g/mol. Therefore, mass (mg) = (0.2 M × 0.0005 L × 1485.71 g/mol) × 1000 = 148.571 mg. Weigh 148.5 mg of vancomycin hydrochloride powder and dissolve in 500 μL of ddH2O. Mix the contents thoroughly until the powder is completely dissolved. Prepare 170 μL aliquots and store them at −20 °C protected from light for a period of 14 days.
- Neurobasal A media containing 0.2X antibiotics (Table 4):
- 1.
- Add 5 mL of the antibiotic cocktail solution to the 500 mL bottle of Neurobasal A media with a sterile serological pipet under sterile conditions to maintain the media sterile.
- 2.
- Mix the contents thoroughly using a 50 mL serological pipet under sterile conditions.
- 3.
- Once all the ingredients are added, securely cap the bottle.
- 4.
- Label the bottle with the date and contents for proper identification.
- 5.
- Store the bottle in a refrigerator at 4 °C until it is used for the preparation of enteric neuron media.
- Glial-Derived Neurotrophic Factor (GDNF) solution (Table 5):
- 1.
- Add 1 mL of sterile ddH2O to the tube containing 10 μg of GDNF from mouse.
- 2.
- Mix the solution: Gently vortex or pipette up and down the contents of the tube to ensure thorough mixing and dissolution of GDNF in the ddH2O. This will result in a solution with a concentration of 10 μg/mL.
- 3.
- Aliquot the solution: Prepare multiple aliquots of the GDNF solution in volumes of 50 μL each. Use sterile pipette tips and microcentrifuge tubes to avoid contamination.
- 4.
- Store at −80 °C
- Enteric neuron media (Table 6):
- 1.
- Add the calculated volume of supplements and solutions to 45.45 mL of Neurobasal A media and using a serological pipet mix the media under sterile conditions to prepare a final volume of 50 mL.
- 2.
- Once all the components are added, securely cap, label, and date the conical tube for proper identification.
- 3.
- Store the bottle in a refrigerator at 4 °C until it is used.
- Fetal Bovine Serum (FBS) is essential for optimal neuronal growth by providing essential nutrients and promoting cell proliferation.
- Glial-Derived Neurotrophic Factor (GDNF) is vital for both neuronal and glial growth, ensuring the development and maintenance of healthy neurons and supporting the growth of glial cells, which play crucial roles in neural function and support.
2.1.2. Small Intestinal Organoid Culture Materials and Reagents
- N-Acetylcysteine: The substance may cause skin and respiratory irritation, and it is highly likely to cause serious eye irritation, warranting warnings for skin, eye, and respiratory effects.
- Ethylenediaminetetraacetic acid (EDTA): H319 indicates a warning for serious eye irritation or damage.
- HA-R-Spondin1-Fc 293T Cells: Caution should be exercised, and the cell line should be handled as potentially bio-hazardous material using Biosafety Level (BSL)-2 containment. Appropriate safety procedures should be used when handling all cell lines. Regular monitoring for mycoplasma contamination is essential to ensure the safety and integrity of cell culture experiments.
- Follow additional steps to prepare crypt culture reagents:
- Preparation of 4 mL solution of 0.1% BSA in PBS 1X (Table 9):
- 1.
- First, prepare a 4 mL solution of 0.1% BSA in PBS 1X.
- 2.
- Weigh 4 mg of BSA and add a small volume of PBS 1X. Ensure that the BSA is fully dissolved.
- 3.
- Add enough PBS 1X to reach a final volume of 4 mL. Mix the solution gently to ensure it is homogeneous.
- 4.
- Filter the solution (pore size of 0.22 μm) and maintain at 4 °C.
- Preparation of 100 μg/mL solution of mNoggin in 0.1% BSA in PBS 1X to create a 1000X stock solution (Table 10):
- 5.
- Take 1 mL of the prepared 0.1% BSA in PBS solution and add it to 100 μg of mNoggin (PreproTech, 250-38).
- 6.
- Mix thoroughly to ensure proper mixing of the components.
- 7.
- Finally, aliquot the solution into 20 μL portions.
- 8.
- Store them at −80 °C for future use.
- Preparation of 500 μg/mL murine Epidermal Growth Factor (mEGF) Recombinant Protein in 0.1% BSA in PBS 1X to create a 10,000× stock solution (Table 11).
- 1.
- Take 2 mL of the prepared 0.1% BSA in PBS 1X solution and add it to 1 mg of Gibco™ mEGF (Fisher Scientific, MG8043).
- 2.
- Mix thoroughly to ensure proper mixing of the components.
- 3.
- Prepare 5 μL aliquots of the solution.
- 4.
- Store the aliquots at −80 °C for future use.
- Preparation of 500 mM N-Acetylcysteine stock solution (Table 12):
- 1.
- Calculate the mass of N-Acetylcysteine powder required to prepare 5 mL at a concentration of 500 mM: Mass (mg) = (0.5 M × 0.005 L × 163.19 g/mol) × 1000 = 407.975 mg.
- 2.
- Weigh 408 mg of N-Acetylcysteine powder (Millipore Sigma, A9165-5G), and dissolve in 5 mL of sterile ddH2O.
- 3.
- Mix thoroughly until the powder is completely dissolved.
- 4.
- Prepare 200 μL aliquot.
- 5.
- Store at −20 °C protected from light.
- N-Acetylcysteine: The substance may cause skin and respiratory irritation, and it is highly likely to cause serious eye irritation, warranting warnings for skin, eye, and respiratory effects.
- Preparation of R-Spondin 1 conditioned media:
- 1.
- R-Spondin 1 secreted protein is expressed by stable transfected 293T cells. These cells are selected using zeocin and express mouse R-spondin 1 protein tagged with hemagglutinin (HA) at the C-terminus and Fc at the N-terminus in the conditioned medium. This cell line was generously provided by the Calvin Kuo Lab at Stanford University and it is now distributed by R&D Systems (Catalog # 3710-001-01). Please refer to R&D Systems (Catalog # 3710-001-01) for protocols to culture, pass, freeze, and collect HA-R-Spondin 1-Fc conditioned media.
- 2.
- Prepare 1 mL aliquots of the collected conditioned media.
- 3.
- Store the aliquots at −20 °C for future use.
- Preparation of supplemented advanced DMEM/F12 media (Table 13):
- 1.
- Add 5 mL of the GlutaMAX, HEPES, and Antibiotic/Antimycotic solution to a bottle of 500 mL Advanced DMEM/F12.
- 2.
- Store at 4 °C.
- 1.
- Add the calculated volume of supplements and solutions to a 16,328 mL supplemented advanced DMEM/F12 media under sterile conditions to prepare a final volume of 20 mL.
- 2.
- Mix thoroughly, securely cap the conical tube, and label appropriately.
- 3.
- Store at 4 °C until use.
- Preparation of EDTA 2 mM in Dulbecco’s phosphate-buffered saline (DPBS) 1X solution (Table 15):
- 4.
- Add 200 μL of the 0.5 M EDTA to the 49.8 mL of DPBS 1X.
- 5.
- Place the solution on ice to keep it cold.
- Prepare DPBS 1X solution with 10% FBS (Table 16):
- 1.
- Add 5 mL of the FBS to the 45 mL of DPBS 1X containing antibiotics and antimycotics to create the DPBS 1X/FBS solution for washing steps.
- 2.
- Place the mixture on ice to keep it cold.
2.2. Procedure
2.2.1. Harvesting of Mouse Small Intestine
- 1.
- C57BL/6J female or male mice (age, 8–12 weeks old) were used for the experiments.
- 2.
- Euthanize the mouse following institutional ethics committee protocols.
- 3.
- Place Krebs solution on ice and bubble it with carbogen (95% oxygen, 5% CO2) for at least 30 min to stabilize pH before tissue harvesting.
- 4.
- Use a single small intestine for the isolation of submucosal and myenteric enteric neurons together with glia cell and small intestinal crypts.
- 5.
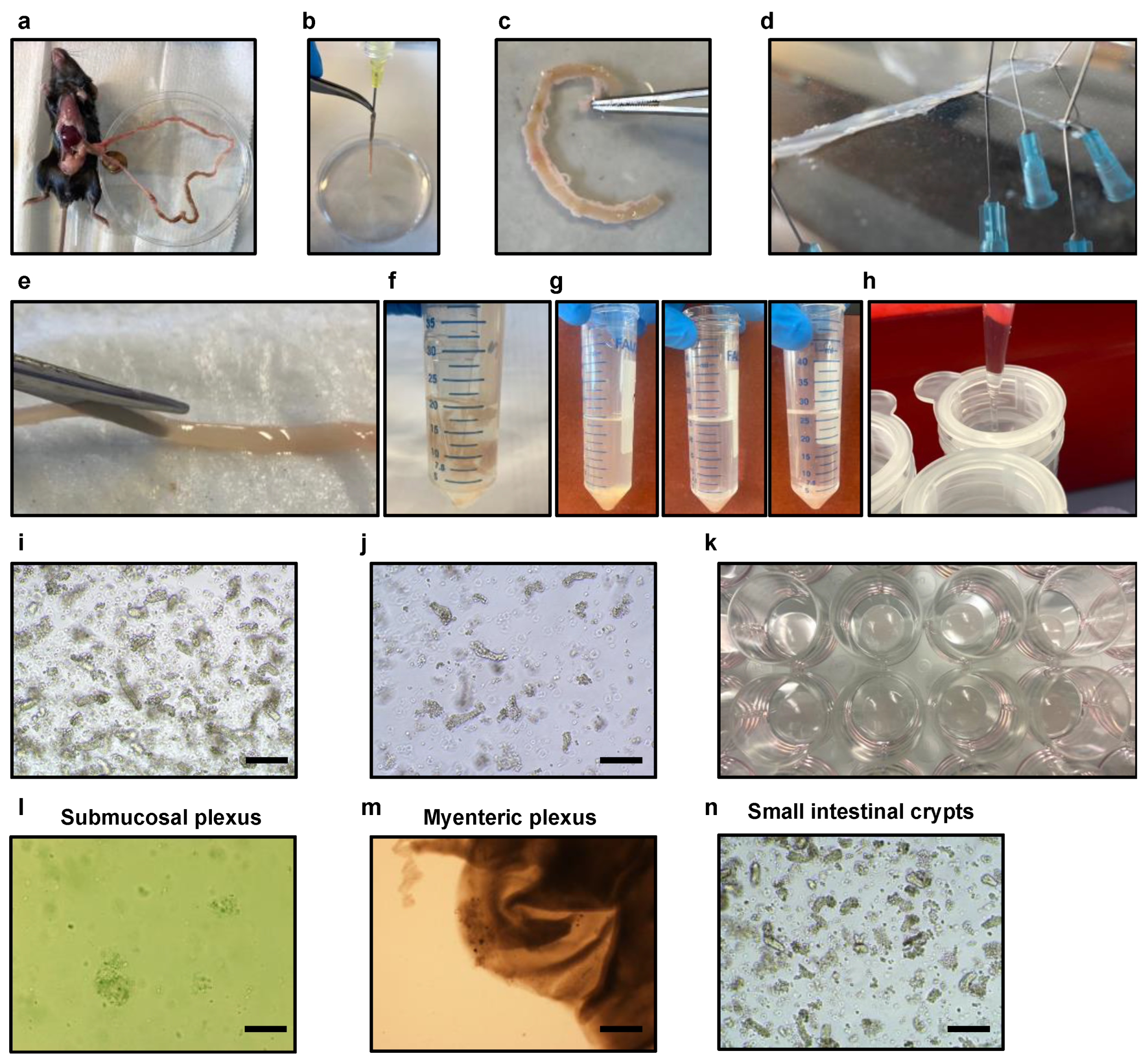
- Prepare the surgical area (abdominal cavity) with ethanol 70%. Perform a small incision with surgical scissors in the epidermis to expose the dermis of the abdominal cavity. Perform another two incisions in the dermis of the abdominal cavity with a clean surgical scissor to expose the small intestine. Harvest the small intestine from the stomach to the cecum of the mice with forceps and clean surgical scissors and place it in a 100 mm cell culture dish containing 10 mL of cold and sterile DPBS 1X supplemented with antibiotics and antimycotics (Figure 2a).
- 6.
- Gently flush small intestinal contents with a 30 mL syringe with a needle (20G) containing sterile and cold DPBS 1X from proximal to distal in a 100 mm cell culture dish until clean (Figure 2b).
- 7.
- Remove any mesenteric tissue, blood vessels, and fat from the exterior of the intestine in a clean 100 mm cell culture dish containing sterile DPBS 1X supplemented with antibiotics and antimycotics (Figure 2c).
- 8.
- Remove and discard the duodenal bulb and the first 2 cm of the duodenum to avoid the Brunner’s glands.
- 9.
- In order to enhance the purity of the isolation procedure, excise the Peyer’s patches along the length of the small intestine.
- 10.
- Divide the small intestine into three parts.
- 11.
- Place the small intestine designated for the isolation of the myenteric and submucosal plexus into a 100 mm cell culture dish covered with 15 mL of ice-cold Krebs solution.
- 12.
- Place the small intestine designated for the isolation of crypts into a 100 mm cell culture dish covered with 15 mL of ice-cold sterile DPBS 1X supplemented with antibiotics and antimycotics.
2.2.2. Isolation of Neurons and Glial Cells from the Myenteric Plexus
- 1.
- Take the small intestine designated for the isolation of the plexuses and place it into a 9 cm silicone-coated black petri dish containing 20–30 mL of ice-chill Kreb’s solution supplemented with 1× antibiotic/antimycotics solution (Figure 2d).
- 2.
- With the use of surgical forceps and a 27G ½ inch needle, secure the proximal and distal parts of the intestine to the silicone (Figure 2d).
- 3.
- Generate a localized region of the myenteric plexus attached to the longitudinal muscle (muscularis externa and serosa) in proximity to the needles by delicately rubbing the outermost layer of the small intestine using a sterile cotton-tipped applicator 6″ 2′s.
- 4.
- Using forceps and a pair of scissors, incise the small intestine along the mesenteric line, carefully exposing the lumen.
- 5.
- Subsequently, remove the needles and turn the small intestine downwards using a pair of angled forceps, ensuring that the lumen side faces down (Figure 2d).
- 6.
- Rearrange the needles to extend the small intestine.
- 7.
- Localize the previously excited muscularis externa with serosa and hold it with a pair of angled forceps.
- 8.
- Using a wet cotton-tipped applicator, gently pull and recover the complete layer of the muscularis externa with serosa along the entire length of the small intestine. Ensure that the forceps are holding the submucosal layer of the intestine down near the site of separation (Figure 2d).
- 9.
- Take a 15 mL conical tube and add 2 mL of ice-cold Krebs solution.
- 10.
- Transfer the isolated muscularis externa with serosa layer to a 100 mm cell culture dish containing an additional 3 mL of Krebs solution.
- 11.
- Using a sterilized razor blade, cut the isolated muscularis externa with serosa layer into the smallest possible pieces.
- 12.
- Using a previously cut and sterile 1 mL pipette tip, collect the resulting 3 mL solution containing the minced muscular externa, and transfer it into the 15 mL conical tube already containing the 2 mL of Krebs solution.
- 13.
- Place it on ice until further use.
2.2.3. Isolation of Neurons and Glial Cells from the Submucosal Plexus
- 1.
- Recover the remaining part of the intestine containing the submucosa and mucosal layer of the intestine by unpinning it from the 9 cm silicone-coated black petri dish.
- 2.
- Rinse it in Krebs solution.
- 3.
- Place the segments into the digestion solution for the submucosal plexus while cutting them into approximately 2 mm pieces with scissors.
- 4.
- Digest it with the prepared digestion solution for the submucosal plexus for 60 min at 37 °C in a water bath with gentle shaking.
- 5.
- Collect the cells by centrifugation for 8 min at 356× g in a refrigerated centrifuge set to 4 °C.
- 6.
- Prepare 0.05% trypsin solution by combining 1 mL of 0.25% trypsin and 4 mL of DPBS 1X supplemented with antibiotics and antimycotics in a sterile 50 mL cell culture tube in the cell culture hood.
- 7.
- Warm the 0.05% trypsin solution at 37 °C in a water bath.
- 8.
- After centrifugation, discard the supernatant without using a vacuum, by decantation into another falcon tube.
- 9.
- Carefully add to the cell pellet 5 mL of 0.05% trypsin solution and mix it gently by pipetting up and down.
- 10.
- Incubate the cells for digestion in a 37 °C water bath for 7 min.
- 11.
- Neutralize the trypsin by adding 500 μL of cold FBS.
- 12.
- Centrifuge the cells for 8 min at 356× g.
- 13.
- While the cells are being centrifuged, place a sterilized 70 μm filter on top of a sterile 15 mL falcon tube.
- 14.
- Remove and discard the supernatant by decantation into another falcon tube.
- 15.
- Gently resuspend the cell mixture by gently pipetting in 5 mL of Krebs solution. Do not generate air bubbles.
- 16.
- Count the cell number using standard Trypan Blue staining and hemocytometry.
- 17.
- Maintain the tube on ice with continuous gentle rotation agitation until subsequent use.
2.2.4. Isolation of Small Intestinal Crypts
- 1.
- Utilizing surgical scissors, cut the intestine into small 2–4 mm fragments. Transfer these fragments into a 50 mL tube containing 20 mL of ice-cold DPBS 1X supplemented with antibiotics and antimycotics (Figure 2f).
- 2.
- Gently and thoroughly cleanse the intestinal fragments by repeatedly pipetting the DPBS 1X solution up and down using a 50 mL serological pipette. Take care to avoid bubble formation during the cleaning process. Allow the intestinal fragments to settle down by gravity, and carefully remove the supernatant containing any debris or impurities. Add fresh ice-cold DPBS 1X supplemented with antibiotics and antimycotics to the tube. Repeat the cleaning process multiple times until the supernatant becomes nearly clear. This requires performing approximately 10 successive washes (Figure 2g).
- 3.
- Remove the supernatant and leave the intestinal fragments in the conical tube.
- 4.
- In this step, 25 mL of a DPBS 1X solution supplemented with 2 mM of EDTA is added to the tube containing the intestinal fragments. EDTA acts as a dissociation agent by chelating divalent cations, such as calcium and magnesium, which are vital for cell-cell adhesion. By disrupting these interactions, EDTA helps loosen the cell-to-cell attachments, facilitating the isolation of single cells from the intestinal epithelium and the isolation of the crypts.
- 5.
- Place the tube on a rocking tube platform within a cold room and allow it to shake for a duration of 30 min to further aid in the dissociation process.
- 6.
- After the incubation period, allow the smaller pieces, primarily consisting of isolated crypts, to undergo gravitational settling.
- 7.
- Once settled, the turbulent supernatant, which contains the single cells and dissociated single cells from the intestinal epithelium, can be carefully removed. This step ensures the separation of the desired crypts from the remaining single cells from the epithelium.
- 8.
- Rapidly add 25 mL of DPBS 1X solution with 10% FBS to the pellet containing the isolated intestinal crypts to provide nutrients and support for the isolated crypt structures. To extract the first fraction of intestinal crypts, the solution is repeatedly pipetted up and down using a 50 mL serological pipette a least 10 times. This process helps release the crypts from the tissue and promotes their collection.
- 9.
- In order to enhance the isolation of crypts and eliminate any residual debris or large clusters, the initial fraction containing the crypts is subjected to filtration utilizing a 70 μm strainer. The tube containing the filtrate is then securely capped and appropriately labeled as the first fraction (Figure 2h).
- 10.
- The isolated crypts are transferred back from the 70 μm strainer into a 50 mL tube, and 25 mL of DPBS 1X solution supplemented with 10% FBS is added. The second fraction of crypts is extracted by gently pipetting the solution up and down using a 50 mL serological pipette for 10 cycles. This process helps release additional crypt structures from the tissue fragments.
- 11.
- The extraction and collection of crypts is repeated for a total of four rounds. After each round, the eluted crypts from the respective fraction are carefully collected and separated. It is important to label each of the four fractions to differentiate and track the isolated crypt populations.
- 12.
- Centrifuge the crypts at 300× g to promote sedimentation.
- 13.
- Resuspend the pellet in 10 mL ice-cold supplemented advanced DMEM/F12 media and proceed with centrifugation at 168× g to remove immune cells. This step helps to enrich the population of intestinal epithelial cells (IECs) by eliminating non-epithelial immune cells.
- 14.
- Resuspend the pellets in 10 mL ice-cold supplemented advanced DMEM/F12 media. This step provides a suitable culture medium to maintain the viability and functionality of the crypts.
- 15.
- Using a pre-wetted pipette tip, take 10 μL of sample from the selected fraction and place it on a glass slide or hemocytometer for precise crypt counting and detailed morphological examination. The purpose of this step is to assess the characteristics and quantity of the crypts.
- 16.
- Utilizing an inverted microscope, carefully examine the size and purity of the crypts, ensuring to avoid fractions that contain a significant number of single cells (Figure 2i,j). Aim to select the fraction or fractions that display a high degree of pure crypts, without substantial contamination by single cells.
- 17.
- Estimate the number of crypts per fraction by performing a specific count that focuses solely on the crypts and excludes single cells. This quantification enables the determination of the crypt population within each fraction accurately.
- 18.
- To create an optimal three-dimensional growth environment for the crypts, calculate and plate a variable range of 100 to 500 crypts, which should be diluted in 30 μL of liquid ice-cold Matrigel.
- 19.
- Carefully place 100–500 crypts, suspended in 30 μL of ice-cold Matrigel, onto a single well of a prewarmed 24-well plate.
2.2.5. Plating the Myenteric Plexus, the Submucosal Plexus, and Isolated Crypts
- 1.
- Thaw aliquots of Matrigel on ice until isolation.
- 2.
- Pre-incubate a 24-well plate in a CO2-incubator.
- 3.
- Plate the dissociated cells into Matrigel, at a density of 1 × 105 enteric neuronal cells derived from the submucosal plexus per 30 μL of Matrigel, which corresponds to one-fifth of a halved small intestine. Therefore, transfer 1 mL of the isolated submucosal plexus (one-fifth) and carefully transfer it to a 15 mL Falcon tube.
- 4.
- For the isolation of crypts, collect the necessary volume to obtain a total of 2600–13,000 crypts, which is equivalent to one-fifth of a halved small intestine. Transfer the collected volume to a 15 mL Falcon tube, ensuring the accurate quantification of the required crypts.
- 5.
- Centrifuge the isolated submucosal, myenteric enteric neurons, and crypts at 356× g for 8 min.
- 6.
- Aspirate and discard all the supernatant using a pipette.
- 7.
- Resuspend the isolated myenteric plexus in 400 μL of ice-cold liquid Matrigel. This step provides a supportive matrix for the myenteric plexus, promoting its preservation and subsequent growth.
- 8.
- Similarly, resuspend the isolated submucosal plexus in 400 μL of ice-cold liquid Matrigel, creating an appropriate environment for the submucosal component.
- 9.
- For the isolated crypts, resuspend them in 780 μL of ice-cold liquid Matrigel, facilitating their embedding within the supportive matrix.
- 10.
- Plate 12 wells of a pre-warmed 24-well plate with 30 μL of Matrigel containing the isolated components of the submucosal plexus (Figure 2k,l and Supplementary Video SA1).
- 11.
- Plate 12 wells of a pre-warmed 24-well plate with 30 μL of Matrigel applied to the center of the well containing the myenteric plexus (Figure 2m).
- 12.
- 13.
- Invert the plates for an additional 30 min in the CO2 cell incubator to promote three-dimensional growth. This inverted position promotes three-dimensional growth, ensuring the crypts, submucosal plexus, and myenteric plexus are appropriately positioned within the Matrigel for their respective growth and development.
- 14.
- Add 500 μL per well of warmed enteric neuron media to the isolated submucosal and myenteric plexuses, providing the necessary nutrient-rich medium for their specific requirements.
- 15.
- Add 500 μL per well of warmed intestinal organoid media to the isolated small intestinal crypts, ensuring the provision of an appropriate medium to support their growth and maintain their cellular characteristics.
2.3. Culture and Expansion of Isolated Enteric Neurons from the Submucosal Plexus, Myenteric Plexus, and Intestinal Crypts
2.4. Optimal Passaging of Small Intestinal Organoids and Medium Change for Enteric Neuronal Cell Cultures after Four Days of Initial Culture
- 1.
- Remove the current culture medium from each well of the 24-well plate using a pipette.
- 2.
- Add 500 μL of cold-supplemented advanced DMEM/F12 media to each well and use a pipette with a bent tip to gently break up the gel and the crypts. Transfer the mixture to a 15 mL tube.
- 3.
- Spin the tube containing the crypts at 600 rpm at 4 °C.
- 4.
- Discard the supernatant.
- 5.
- Add 780 μL of ice-cold liquid Matrigel into the tube, which is equivalent to 30 μL per well. To further fragment the intestinal organoids, employ a pre-wetted pipette with a bent tip, and perform pipetting up and down motions.
- 6.
- Position droplets containing 30 μL of crypts embedded in Matrigel at the center of each pre-warmed well in the 24-well plate.
- 7.
- Add 500 μL of complete small intestinal growth media to facilitate proper small intestinal growth.
- 8.
- Transfer the plate to a CO2 incubator at 37 °C.
- 9.
- Allow the cells to grow for an additional four days.
2.5. Staining Procedures
3. Results
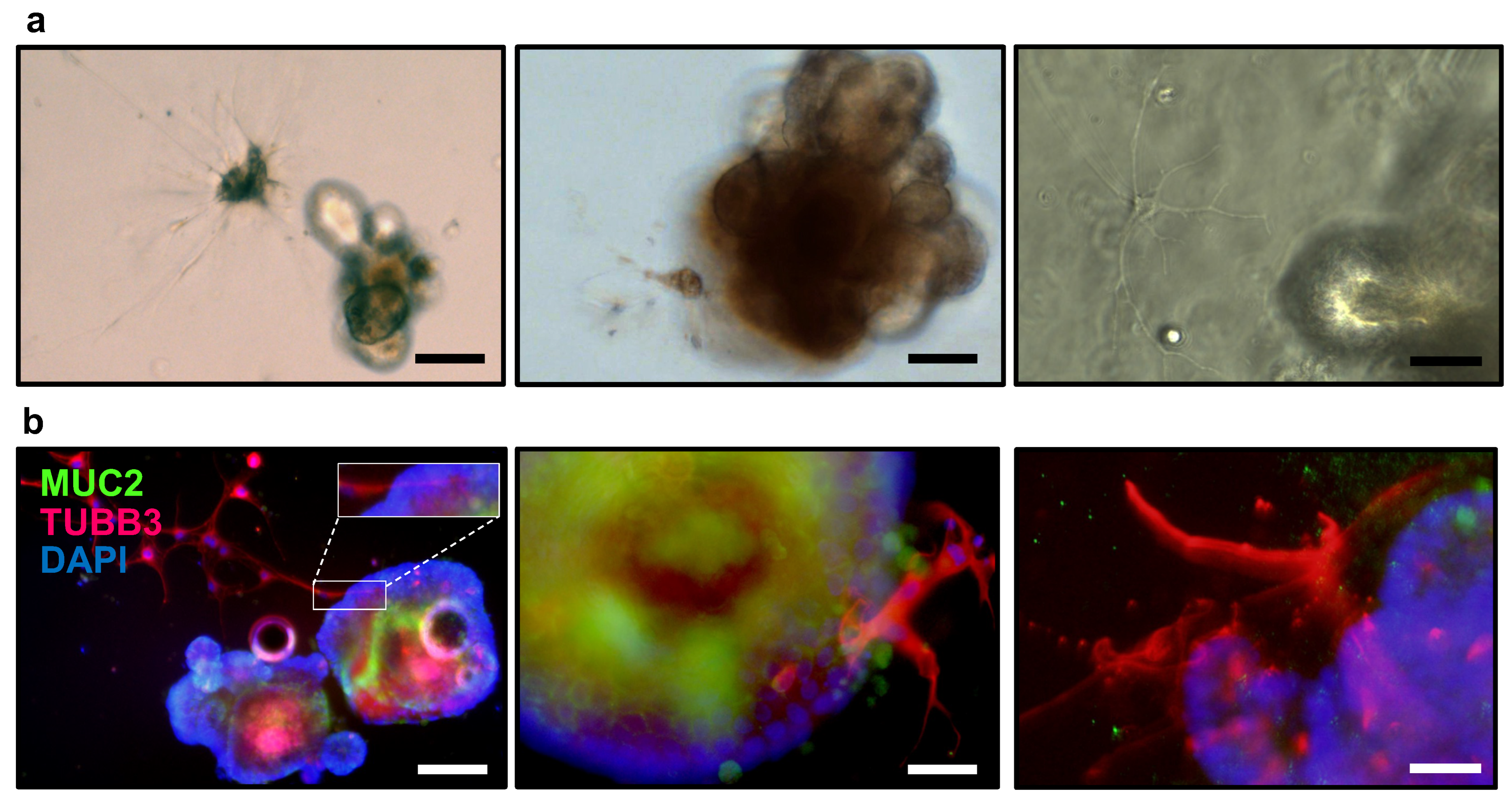
3.1. Co-Culture of Small Intestinal Organoids and Isolated Enteric Neurons from Submucosal and Myenteric Plexus
- 1.
- Warm the 0.25% trypsin solution at 37 °C in a water bath.
- 2.
- Remove the current neuronal culture medium from each well of the 24-well plate using a pipette.
- 3.
- Add 500 μL of cold supplemented advanced DMEM/F12 media to each well and use a pipette with a bent tip to gently break up the gel and the crypts. Transfer the mixture to a 15 mL tube.
- 4.
- Spin the tube containing the crypts at 600 rpm at 4 °C.
- 5.
- Discard the supernatant.
- 6.
- Add 3 mL of warm 0.25% trypsin solution to the pellet.
- 7.
- Incubate the cells for digestion in a 37 °C water bath for 7 min.
- 8.
- Neutralize the trypsin by adding 500 μL of cold FBS.
- 9.
- Centrifuge the cells for 8 min at 356× g.
- 10.
- While the cells are being centrifuged, place a sterilized 70 μm filter on top of a sterile 15 mL falcon tube.
- 11.
- Remove and discard the supernatant.
- 12.
- Gently resuspend the cell mixture by gently pipetting in 5 mL of DMEM/F12 media. Do not generate air bubbles.
- 13.
- Count the cell number using standard Trypan Blue staining and hemocytometry.
- 14.
- Calculate to seed at a density of 1.5 × 104 enteric neurons from the myenteric plexus per 30 μL of Matrigel.
- 15.
- Calculate to seed at a density of 2.5 × 104 enteric neurons from the submucosal plexus per 30 μL of Matrigel. Note the submucosal neurons are generally smaller in size compared to myenteric neurons due to functional specialization.
- 16.
- We are preparing 48 wells so therefore we collect the appropriate volume to gather 7.2 × 105 enteric neurons from the myenteric plexus and 1.2 × 106 isolated enteric neurons from the submucosal plexus.
- 17.
- Centrifuge the neuronal cells for 8 min at 356× g.
- 18.
- Remove and discard the supernatant.
- 19.
- Add 390 μL of ice-cold liquid Matrigel to 7.2 × 105 isolated enteric neurons from the myenteric plexus.
- 20.
- Add 390 μL of ice-cold liquid Matrigel to 1.2 × 106 isolated enteric neurons from the submucosal plexus.
- 21.
- Keep on ice.
- Calculate to see crypts at a density of 150 crypts in 30 μL of liquid ice-cold Matrigel. We are plating 48 wells so 7200 crypts are diluted in 750 μL of ice-cold liquid Matrigel.
- Keep on ice.
- 1.
- Combine 390 μL of ice-cold liquid Matrigel containing 7.2 × 105 isolated enteric neurons from the myenteric plexus, 390 μL of ice-cold liquid Matrigel containing 1.2 × 106 isolated enteric neurons from the submucosal plexus, and 750 μL of ice-cold liquid Matrigel containing 7200 crypts.
- 2.
- With a pre-wetted pipette perform pipetting up and down motions for proper mixture.
- 3.
- Position droplets containing 30 μL of crypts embedded in Matrigel at the center of each pre-warmed well in the 24-well plate. Plate 48 wells. If immunofluorescence or immunohistochemical analysis is desired, plating should be performed on a 4-well chamber slide (Thermo Scientific, 154526).
- 4.
- Add 500 μL of complete small intestinal growth media to facilitate proper growth and innervations.
- 5.
- Transfer the plate to a CO2 incubator at 37 °C.
- 6.
- Allow the cells to grow for an additional four days.
3.2. Microinjection of Fluorescently Labeled Conjugates into Small Intestinal Organoids: A Versatile Technique for Investigating Cellular Integrity, Intestinal Permeability, and Neuronal-Epithelial Interactions
- 1.
- Prepare the microinjector according to the manufacturer’s instructions.
- 2.
- Position the microinjector near the microscope on a stable platform.
- 3.
- Use the micropipette puller to pull glass capillaries (TW100F-6, WPI, with a 1.0 mm outer diameter and 0.75 mm bore) under the following conditions: heat (400), filament (4), velocity (50), delay (250), pull (200). Aim for an inner diameter of approximately 15 μm at the tip of the capillary. Pull glass capillaries to a length of 5.5 cm. Ensure the glass capillary needles are sterilized.
- 4.
- Fill the glass capillary needle with mineral oil using a long steel needle.
- 5.
- Attach the capillary to the microinjector, ensuring that there is no air trapped inside by emptying the mineral oil from the capillary.
- 6.
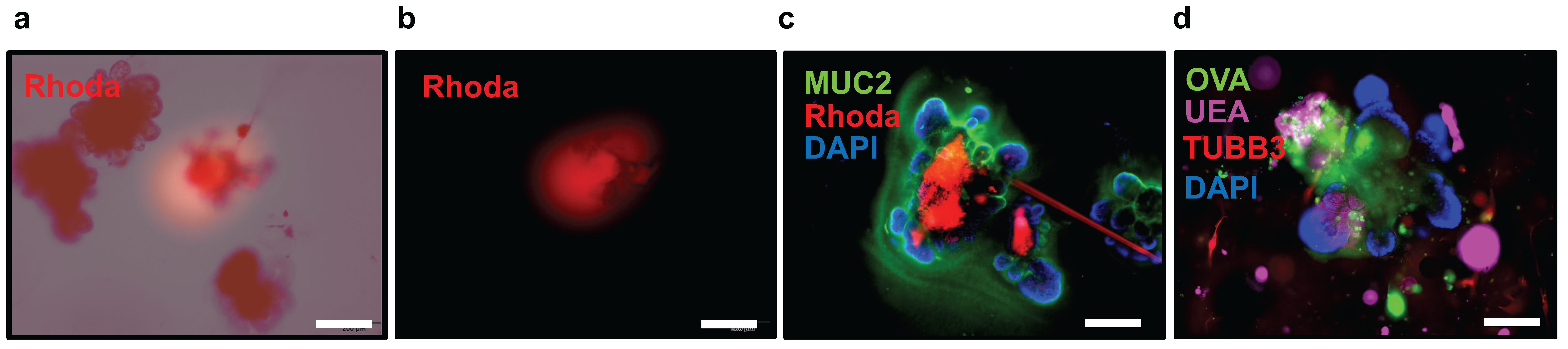
- Prepare a small chamber and add fluorescently labeled dextran or ovalbumin to one of the wells. Fill the capillary with 50 μL of the fluorescently labeled compound, taking care to avoid any breakage of the capillary tip.
- 7.
- Move the co-culture of enteric neurons and the organoids chamber from the cell culture incubator to a microscope set up in close proximity to the microinjector.
- 8.
- Open the chamber lid and adjust the micromanipulator to position the capillary tip above the first organoid, just above the culture media. Use the Z-axis control of the micromanipulator to advance the capillary, first penetrating the Matrigel until reaching the organoid surface.
- 9.
- Carefully penetrate the organoid with the microcapillary tip and inject 50 nL of the fluorescently labeled compound.
- 10.
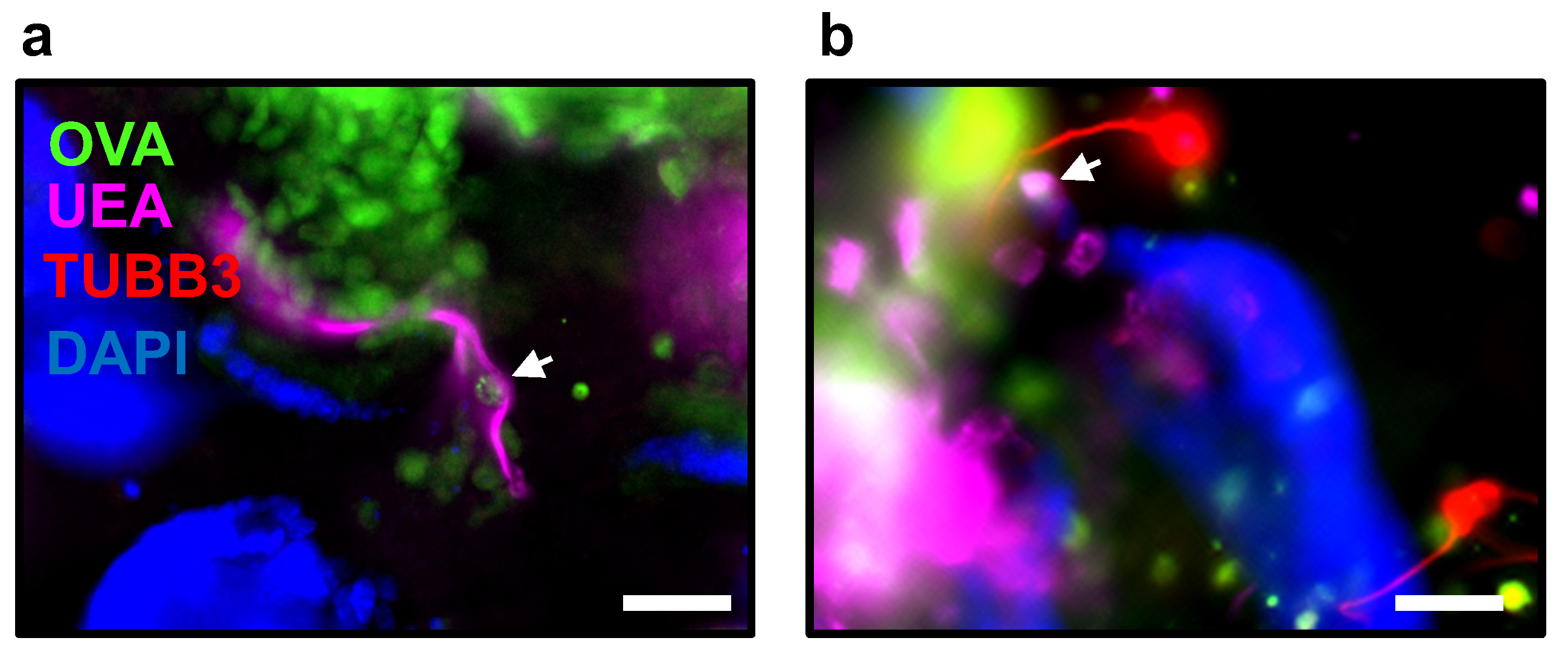
- Monitor the microinjection process by activating the fluorescence light source to excite the fluorophores and observe the emitted light from the fluorescently labeled compound within the organoid (Figure 6a,b).
- 11.
- After injection, retract the needle to the media surface and proceed to the next organoid for microinjection.
- 12.
- Carry out the subsequent staining procedures.
4. Limitations
5. Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Almeqdadi, M.; Mana, M.D.; Roper, J.; Yilmaz, O.H. Gut organoids: Mini-tissues in culture to study intestinal physiology and disease. Am. J. Physiol. Cell Physiol. 2019, 317, C405–C419. [Google Scholar] [CrossRef] [PubMed]
- Amen, A.M.; Ruiz-Garzon, C.R.; Shi, J.; Subramanian, M.; Pham, D.L.; Meffert, M.K. A Rapid Induction Mechanism for Lin28a in Trophic Responses. Mol. Cell 2017, 65, 490–503.e497. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.F.; Doherty, D.H.; Lile, J.D.; Bektesh, S.; Collins, F. GDNF: A glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 1993, 260, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Fairman, W.A.; Vandenberg, R.J.; Arriza, J.L.; Kavanaugh, M.P.; Amara, S.G. An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature 1995, 375, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Lima, M.M.; Procopio, J.; Pithon-Curi, T.C.; Doi, S.Q.; Bazotte, R.B.; Curi, R. Glutamine and glutamate as vital metabolites. Braz. J. Med. Biol. Res. 2003, 36, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Haramis, A.P.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Date, S.; Sato, T. Mini-gut organoids: Reconstitution of the stem cell niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef]
- Frey, M.R.; Golovin, A.; Polk, D.B. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J. Biol. Chem. 2004, 279, 44513–44521. [Google Scholar] [CrossRef]
- Suzuki, A.; Sekiya, S.; Gunshima, E.; Fujii, S.; Taniguchi, H. EGF signaling activates proliferation and blocks apoptosis of mouse and human intestinal stem/progenitor cells in long-term monolayer cell culture. Lab. Investig. 2010, 90, 1425–1436. [Google Scholar] [CrossRef]
- de Lau, W.; Peng, W.C.; Gros, P.; Clevers, H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 2014, 28, 305–316. [Google Scholar] [CrossRef]
- Hamnett, R.; Dershowitz, L.B.; Sampathkumar, V.; Wang, Z.; Gomez-Frittelli, J.; De Andrade, V.; Kasthuri, N.; Druckmann, S.; Kaltschmidt, J.A. Regional cytoarchitecture of the adult and developing mouse enteric nervous system. Curr. Biol. 2022, 32, 4483–4492.e4485. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Bixby, S.; Kruger, G.M.; Mosher, J.T.; Joseph, N.M.; Morrison, S.J. Cell-intrinsic differences between stem cells from different regions of the peripheral nervous system regulate the generation of neural diversity. Neuron 2002, 35, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Zakhem, E.; Rego, S.L.; Raghavan, S.; Bitar, K.N. The appendix as a viable source of neural progenitor cells to functionally innervate bioengineered gastrointestinal smooth muscle tissues. Stem Cells Transl. Med. 2015, 4, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Kriegstein, A.; Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009, 32, 149–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Llorente, C.; Hartmann, P.; Yang, A.M.; Chen, P.; Schnabl, B. Methods to determine intestinal permeability and bacterial translocation during liver disease. J. Immunol. Methods 2015, 421, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, X.; Andersson, R. Role of intestinal permeability in monitoring mucosal barrier function. History, methodology, and significance of pathophysiology. Dig. Surg. 1998, 15, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Duffey, M.E.; Hainau, B.; Ho, S.; Bentzel, C.J. Regulation of epithelial tight junction permeability by cyclic AMP. Nature 1981, 294, 451–453. [Google Scholar] [CrossRef]
- Simon, D.B.; Lu, Y.; Choate, K.A.; Velazquez, H.; Al-Sabban, E.; Praga, M.; Casari, G.; Bettinelli, A.; Colussi, G.; Rodriguez-Soriano, J.; et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999, 285, 103–106. [Google Scholar] [CrossRef]
- Hollander, D. Crohn’s disease—A permeability disorder of the tight junction? Gut 1988, 29, 1621–1624. [Google Scholar] [CrossRef]
- Katz, K.D.; Hollander, D.; Vadheim, C.M.; McElree, C.; Delahunty, T.; Dadufalza, V.D.; Krugliak, P.; Rotter, J.I. Intestinal permeability in patients with Crohn’s disease and their healthy relatives. Gastroenterology 1989, 97, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.; Vogelsang, H.; Hubl, W.; Waldhoer, T.; Lochs, H. Intestinal permeability and the prediction of relapse in Crohn’s disease. Lancet 1993, 341, 1437–1439. [Google Scholar] [CrossRef] [PubMed]
- Llorente, C.; Schnabl, B. The gut microbiota and liver disease. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Mande, S.S. Host-microbiome interactions: Gut-Liver axis and its connection with other organs. NPJ Biofilms Microbiomes 2022, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- McDole, J.R.; Wheeler, L.W.; McDonald, K.G.; Wang, B.; Konjufca, V.; Knoop, K.A.; Newberry, R.D.; Miller, M.J. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012, 483, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Knoop, K.A.; McDonald, K.G.; McCrate, S.; McDole, J.R.; Newberry, R.D. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 2015, 8, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Bruellman, R.; Llorente, C. A Perspective Of Intestinal Immune-Microbiome Interactions In Alcohol-Associated Liver Disease. Int. J. Biol. Sci. 2021, 17, 307–327. [Google Scholar] [CrossRef]
- Burns, A.J.; Goldstein, A.M.; Newgreen, D.F.; Stamp, L.; Schafer, K.H.; Metzger, M.; Hotta, R.; Young, H.M.; Andrews, P.W.; Thapar, N.; et al. White paper on guidelines concerning enteric nervous system stem cell therapy for enteric neuropathies. Dev. Biol. 2016, 417, 229–251. [Google Scholar] [CrossRef]
- Workman, M.J.; Mahe, M.M.; Trisno, S.; Poling, H.M.; Watson, C.L.; Sundaram, N.; Chang, C.F.; Schiesser, J.; Aubert, P.; Stanley, E.G.; et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017, 23, 49–59. [Google Scholar] [CrossRef]
- Levin, D.E.; Mandal, A.; Fleming, M.A.; Bae, K.H.; Gerry, B.; Moore, S.R. Intestinal crypt-derived enteroid coculture in presence of peristaltic longitudinal muscle myenteric plexus. Biol. Methods Protoc. 2021, 6, bpaa027. [Google Scholar] [CrossRef]
- Fattahi, F.; Steinbeck, J.A.; Kriks, S.; Tchieu, J.; Zimmer, B.; Kishinevsky, S.; Zeltner, N.; Mica, Y.; El-Nachef, W.; Zhao, H.; et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 2016, 531, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Medina, I.; Jevans, B.; Boismoreau, F.; Chettouh, Z.; Enomoto, H.; Muller, T.; Birchmeier, C.; Burns, A.J.; Brunet, J.F. Dual origin of enteric neurons in vagal Schwann cell precursors and the sympathetic neural crest. Proc. Natl. Acad. Sci. USA 2017, 114, 11980–11985. [Google Scholar] [CrossRef]
- Okamoto, R.; Shimizu, H.; Suzuki, K.; Kawamoto, A.; Takahashi, J.; Kawai, M.; Nagata, S.; Hiraguri, Y.; Takeoka, S.; Sugihara, H.Y.; et al. Organoid-based regenerative medicine for inflammatory bowel disease. Regen. Ther. 2020, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordonez-Moran, P.; Clevers, H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Kim, H.J. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat. Protoc. 2022, 17, 910–939. [Google Scholar] [CrossRef] [PubMed]
- Roper, J.; Tammela, T.; Akkad, A.; Almeqdadi, M.; Santos, S.B.; Jacks, T.; Yilmaz, O.H. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 2018, 13, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kobayashi, S.; Ogasawara, N.; Okamoto, R.; Nakamura, T.; Watanabe, M.; Jensen, K.B.; Yui, S. Transplantation of intestinal organoids into a mouse model of colitis. Nat. Protoc. 2022, 17, 649–671. [Google Scholar] [CrossRef]
- Fumagalli, A.; Suijkerbuijk, S.J.E.; Begthel, H.; Beerling, E.; Oost, K.C.; Snippert, H.J.; van Rheenen, J.; Drost, J. A surgical orthotopic organoid transplantation approach in mice to visualize and study colorectal cancer progression. Nat. Protoc. 2018, 13, 235–247. [Google Scholar] [CrossRef]



| Vf = 1 L | mg | Cf (mM) |
|---|---|---|
| Sodium chloride (NaCl) (Millipore Sigma, Burlington, MA, USA, S5886) | 6895 | 118 |
| Potasium chloride (KCl) (Millipore Sigma, P9333) | 343 | 4.6 |
| Sodium dihydrogen phosphate (NaH2PO4) (Millipore Sigma, 1.06370) | 156 | 1.3 |
| Magnesium sulfate (MgSO4) (Millipore Sigma, 746452) | 144.5 | 1.2 |
| Sodium bicarbonate (NaHCO3) (Millipore Sigma, S5761) | 2100 | 25 |
| Glucose (Millipore Sigma, D9434) | 1980 | 11 |
| Calcium chloride (CaCl2) (Millipore Sigma, C4901) | 277.5 | 2.5 |
| Gibco™ Antibiotic/Antimycotic, 100X (ThermoFisher, Waltham, MA, USA, 15240062), −20 °C | 10 mL | 1X |
| Vf = 10 mL | Cf (mM) | |
|---|---|---|
| Collagenase, Type II, 1 g (ThermoFisher, 17101015) | 13 mg | 1.3 mg/mL |
| Bovine Serum Albumin Fraction V, heat shock from bovine serum, 250 g, (Roche, Basel, Switzerland, 3116956001), 4 °C | 3 mg | 0.3 mg/mL |
| Carbogen-bubbled Krebs solution | 10 mL |
| Vf = 10 mL | Vo (μL) | Cf |
|---|---|---|
| Ampicillin sodium solution 100 mg/mL (Goldbio, St. Louis, MO, USA, A-301-25), −20 °C | 400 | 0.2 mg/mL |
| Gentamycin, 10 mg/mL (Millipore Sigma, G1272-10ML), 4 °C | 1000 | 0.05 mg/mL |
| Gibco™ Antibiotic/Antimycotic, 100× (ThermoFisher, 15240062), −20 °C | 2000 | 20X |
| Neomycin, 10 mg/mL (Millipore Sigma, N1142-20ML), 4 °C | 1000 | 0.05 mg/mL |
| Metronidazole, 50 mg/mL (Millipore Sigma, M3761-5G), 4 °C | 1000 | 0.25 mM |
| Vancomycin hydrochloride solution 200 mM (Millipore Sigma, 1709007-500MG), −20 °C | 160 | 0.16 mM |
| Vf = 500 mL | mL | Cf |
|---|---|---|
| Antibiotic cocktail 20X, −20 °C | 5 | 0.2X |
| Neurobasal™-A Medium, minus phenol red (Gibco, 12349015), 4 °C | 500 |
| Vf = 50 μL | Cf | |
|---|---|---|
| Glial-Derived Neurotrophic Factor (GDNF) (Millipore Sigma, SRP3200), −80 °C | 10 μg | 10 μg/mL |
| ddH2O | 1 mL |
| Vf = 50 mL | Vo (μL) | Cf |
|---|---|---|
| Gibco™ Sodium pyruvate, 100 mM (ThermoFisher, 11360070) | 500 | 1 mM |
| Gibco™ GlutaMAX™ supplement, 100 mL, 100X (ThermoFisher, 35050061) | 500 | 2 |
| Gibco™ B-27™ supplement, 50X (ThermoFisher,17504044) | 1000 | 1X |
| Glial-Derived Neurotrophic Factor (GDNF) solution, 10 μg/mL | 50 | 0.01 μg/mL |
| Fetal Bovine Serum (FBS) heat inactivated (Omega Scientific, FB-02) | 5000 | 10% |
| Antibiotic cocktail (20X) | 2500 | 1X |
| Neurobasal A media containing 0.2X antibiotics | 40,450 |
| Falcon 50 mL Conical Centrifuge Tubes (Corning, Corning, NY, USA, 1495949A) |
| Falcon 15 mL Conical Centrifuge Tubes (Fisher Scientific, 14-959-49B) |
| Biopioneer 50 mL Serological Pipet, 100/pack (Fisher Scientific, GEX500-S01) |
| Biopioneer 25 mL Serological Pipet 200/pack (Fisher Scientific, GEX250S01) |
| Biopioneer 10 mL Serological Pipet 200/pack (Fisher Scientific, GEX0100-S01) |
| Surgical forceps and scissors |
| Corning™ Disposable Vacuum Filter/Storage Systems 500 mL, 0.2 μm (Fisher Scientific, 430769) |
| Falcon® 70 µm Cell Strainer, White, Sterile (Corning, 352350) |
| Whatman Puradisc 25 mm PES Syringe Filters, 0.2 µm, 50 pack, (50 units) (Whatman, Maidstone, UK, 6780-2502) |
| Cotton Tip Applicator 6″ 2′s Sterile 100/box (Dinarex, Montvale, NJ, USA, 4305) |
| Falcon® 150 mm Tissue Culture -treated Cell Culture Dish with 20 mm Grid, 10/Pack, 100/Case, Sterile (Corning 353025) |
| Falcon® 100 mm Tissue Culture-treated Cell Culture Dish, 20/Pack, 200/Case, Sterile (Corning, 353003) |
| Falcon® 24-well Clear Flat Bottom Tissue Culture-treated Multiwell Cell Culture Plate, with Lid, Sterile, 50/Case, (Corning, 353047) |
| Syringe with BD Luer-Lok™ Tip, 30 mL (BD302832, VWR) |
| Syringe 27Gx1/2″ (BD#309623) 1 mL syringe, (Fisher Scientific, 1482687) |
| 25G 5/8 Needle (BD# 305122), (Fisher Scientific, 14-826AA) |
| BD® Needle 1/2 in. single use, sterile, 27 G (BD, 305109) |
| BD Precisionglide® Needle 21G × 1 (0.8 mm × 25 mm), (BD, 305165) |
| Dissection Dish, Large, Black 3-Pack (Living systems instrumentations, DD-90-S-BLK-3PK) |
| Fisherbrand™ Razor Blades,100/pk, (Fisher Scientific, 12-640) |
| Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, 10 mL (Millipore Sigma, CLS356231). Prepare 1 mL aliquots and store at −80 °C |
| Advanced DMEM/F12, 500 mL (ThermoFisher 12634010), 4 °C |
| Gibco™ GlutaMAX™ supplement (ThermoFisher, 35050061), 4 °C |
| Gibco™ (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid) (HEPES) 1 M, 100 mL, (ThermoFisher, 15630080), 4 °C |
| Gibco™ Antibiotic/Antimycotic, 100X (ThermoFisher, 15240062), −20 °C |
| Gibco™ B-27™ supplement, 50X, 10 mL (ThermoFisher, 17504044). Prepare 400 μL aliquots and stored at −20 °C |
| Gibco™ N2 supplement, 100X, 5 mL (ThermoFisher, 17502048). Prepare 200 μL aliquots and store at −20 °C |
| Recombinant murine Noggin (mNoggin), 100 ug, (Peprotech, Cranbury, NJ, USA, 250-38). Prepare 100 μg/mL solution of mNoggin in 0.1% Bovine Serum Albumin (BSA) in Phosphate-Buffered Saline (PBS) to create a 1000X stock solution |
| 1 mg of Gibco™ murine Epidermal Growth Factor (mEGF) Recombinant Protein (MG8043, fisherscientific). Prepare 500 μg/mL murine in 0.1% Bovine Serum Albumin (BSA) in Phosphate-buffered saline (PBS) to create a 10,000X stock solution. Store at −80 °C. |
| Fetal Bovine Serum (FBS) heat inactivated (Omega Scientific, Singapore, FB-02). Aliquot in 50 mL and store at −20 °C |
| N-Acetylcysteine, 5 G, (Millipore Sigma, A9165-5G), 4 °C. Prepare a 500 mM stock solution. |
| Gibco™ DPBS, no calcium, no magnesium (ThermoFisher, 14190136). Supplement with 5 mL of GibcoTM Antibiotic/Antimycotic, 100× (ThermoFisher, 15240062). 4 °C. |
| Ethylenediaminetetraacetic acid (EDTA) (0.5 M), pH 8.0, RNase-free (ThermoFisher, AM9260G), 4 °C |
| R-Spondin 1 conditioned media by a stable transfected 293T cell selected by zeocin expressing mouse R-spondin1 protein tagged with C-terminus HA and N-terminus Fc in the conditioned medium. (Kind gift from Calvin Kuo lab at Stanford University) (R&D Systems, Minneapolis, MN, USA, 3710-001-01). |
| Vf = 4 mL | Cf | |
|---|---|---|
| Bovine Serum Albumin Fraction V, heat shock from bovine serum, 250 g, (Roche, 3116956001), 4 °C | 4 mg | 0.1% |
| Phosphate Buffered Saline (PBS), 492 g, pH 7.4 (Fisher BioReagents, Pittsburgh, PA, USA, BP661-50). Prepare PBS 1X by mixing 9.84 g of PBS in 1 L of ddH2O and sterilize by autoclaving or filter sterilization, 4 °C. | 4 mL |
| Vf = 1 mL | Cf | |
|---|---|---|
| mNoggin, 100 μg, (PeproTech, 250-38), −80 °C | 100 μg | 100 μg/mL |
| 0.1% Bovine Serum Albumin (BSA) in Phosphate-buffered saline (PBS), 4 °C | 1 mL |
| Vf = 2 mL | Cf | |
|---|---|---|
| 1 mg of Gibco™ Epidermal Growth Factor (mEGF) Recombinant Protein (PMG8043, Fisher Scientific), −80 °C | 1 mg | 500 μg/mL |
| 0.1% Bovine Serum Albumin (BSA) in Phosphate-Buffered Saline (PBS) 1X | 2 mL |
| Vf = 5 mL | Cf | |
|---|---|---|
| N-Acetylcysteine, 5 G, 163.19 g/mol, (Millipore Sigma, A9165-5G), 4 °C. | 1 mg | 500 mM |
| Sterile ddH2O. | 5 mL |
| Vf = 500 mL | Vo (mL) | Cf |
|---|---|---|
| Advanced DMEM/F12, 500 mL (ThermoFisher, 12634010), 4 °C | 500 | |
| Gibco™ GlutaMAX™ supplement, 100 mL, 100X (ThermoFisher, 35050061), 4 °C | 5 | 1X |
| Gibco™ (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid) (HEPES) 1 M, 100 mL, (ThermoFisher 15630080), 4 °C | 5 | 10 mM |
| Gibco™ Antibiotic/Antimycotic, 100X (ThermoFisher, 15240062), −20 °C | 5 | 1X |
| Vf = 20 mL | Vo | Cf |
|---|---|---|
| Gibco™ B-27™ supplement, 50X, 10 mL (ThermoFisher, 17504044), −20 °C | 400 μL | 1X |
| Gibco™ N2 supplement, 100X, 5 mL (ThermoFisher, 17502048), −20 °C | 200 μL | 1X |
| 500 mM N-Acetylcysteine stock solution, −20 °C | 50 μL | 10 mM |
| R-Spondin 1 conditioned media, −20 °C | 2000 μL | 1X |
| 500 μg/mL mEGF, 10,000X stock solution, −80 °C | 2 μL | 0.05 μg/mL |
| 100 μg/mL mNoggin, 1000X stock solution, −80 °C | 20 μL | 0.1 μg/mL |
| Antibiotic cocktail (20X), −20 °C | 1000 μL | 1X |
| Supplemented advanced DMEM/F12, 4 °C | 16,328 μL |
| Vf = 25 mL | Vo | Cf |
|---|---|---|
| EDTA (0.5 M), pH 8.0, RNase-free (ThermoFisher, AM9260G), 4 °C | 100 μL | 2 mM |
| Gibco™ DPBS, no calcium, no magnesium (ThermoFisher, 14190136) supplemented with 5 mL of GibcoTM Antibiotic/Antimycotic, 100× (ThermoFisher, 15240062). 4 °C. | 24.9 mL |
| Vf = 100 mL | Vo (mL) | Cf |
|---|---|---|
| Fetal Bovine Serum (FBS) heat inactivated (Omega Scientific, FB-02), 4 °C | 10 | 10% |
| Gibco™ DPBS, no calcium, no magnesium (ThermoFisher, 14190136) supplemented with 5 mL of GibcoTM Antibiotic/Antimycotic, 100× (ThermoFisher, 15240062), 4 °C | 90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llorente, C. Isolation of Myenteric and Submucosal Plexus from Mouse Gastrointestinal Tract and Subsequent Co-Culture with Small Intestinal Organoids. Cells 2024, 13, 815. https://doi.org/10.3390/cells13100815
Llorente C. Isolation of Myenteric and Submucosal Plexus from Mouse Gastrointestinal Tract and Subsequent Co-Culture with Small Intestinal Organoids. Cells. 2024; 13(10):815. https://doi.org/10.3390/cells13100815
Chicago/Turabian StyleLlorente, Cristina. 2024. "Isolation of Myenteric and Submucosal Plexus from Mouse Gastrointestinal Tract and Subsequent Co-Culture with Small Intestinal Organoids" Cells 13, no. 10: 815. https://doi.org/10.3390/cells13100815





