Exploring the Regulators of Keratinization: Role of BMP-2 in Oral Mucosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Laser Microdissection (LMD)
2.3. Libraries Preparation and Sequencing
2.4. Bioinformatic Analysis of Bulk RNA-Seq Data
2.5. Cell Culture
2.6. RT-qPCR
2.7. Histological Analysis
2.8. Statistical Analysis
3. Results
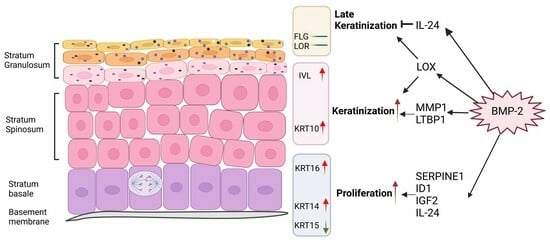
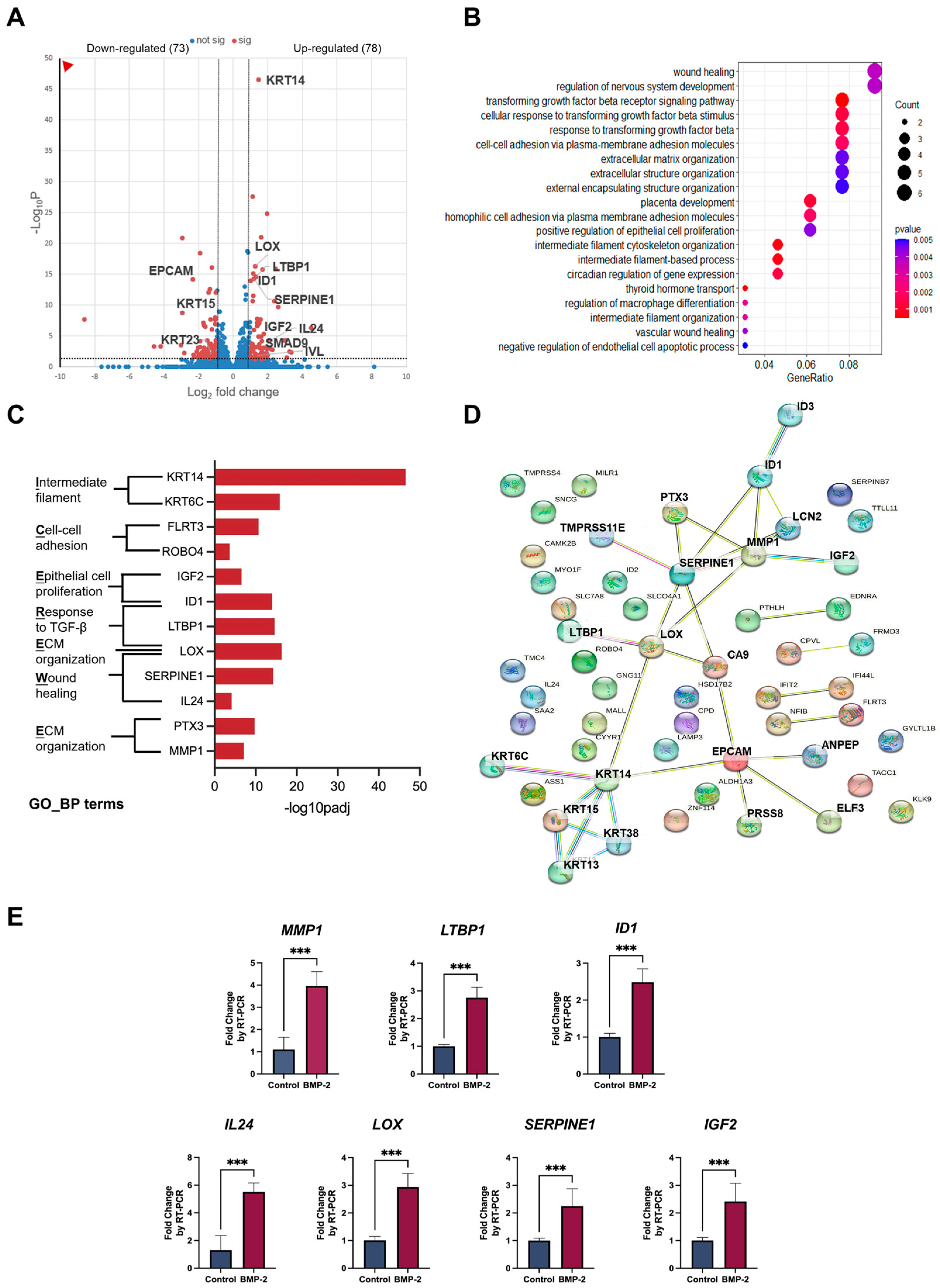
3.1. Extracellular Matrix and Ossification-Associated Genes Were Candidate Regulators of Keratinization
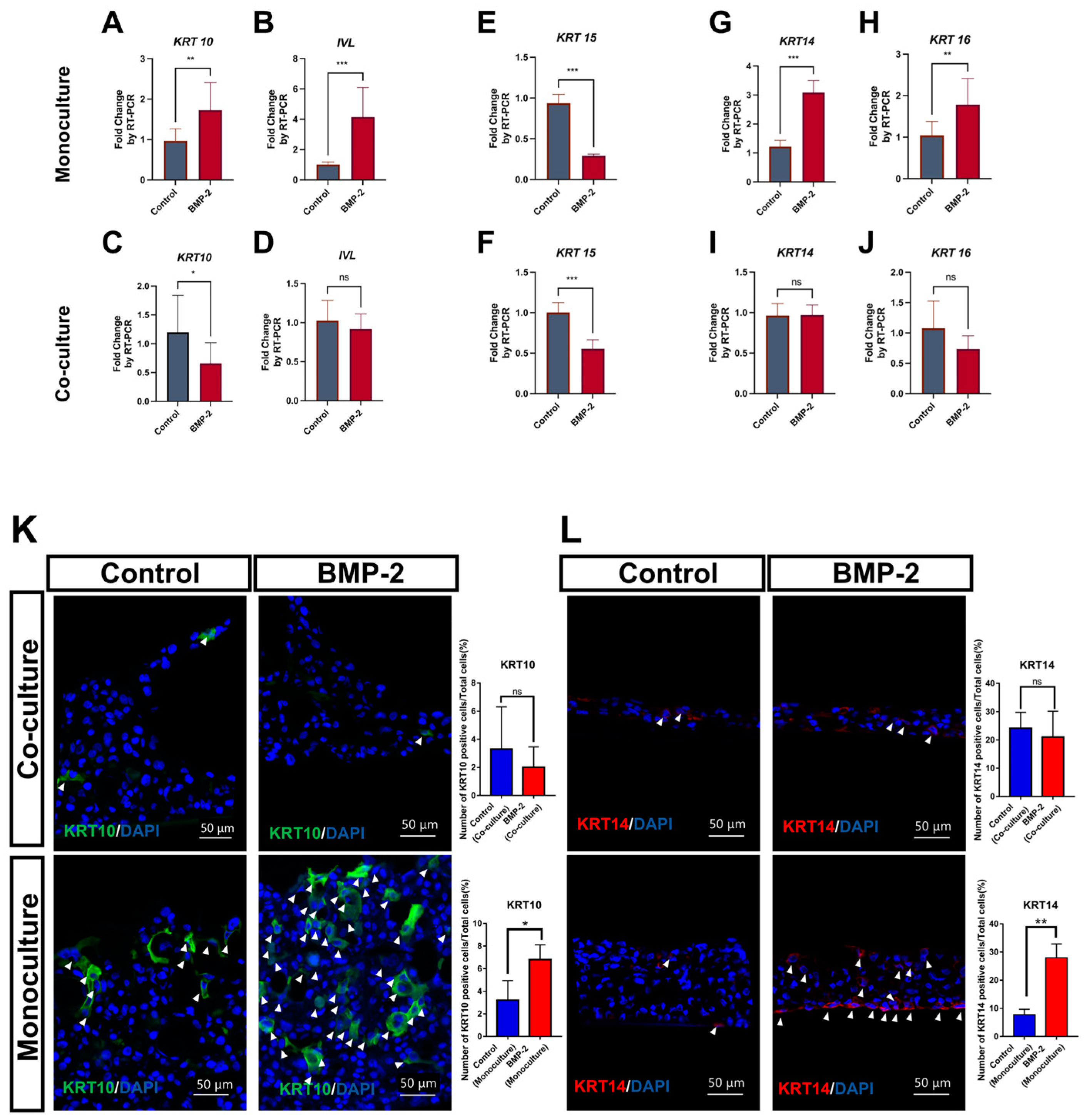
3.2. BMP-2 Functions as an Inducer of Keratinization and Proliferation in ALI Culture
3.3. The Downstream Targets of BMP-2 Are Crucial in Regulating the Keratinization of Oral Mucosa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groeger, S.; Meyle, J. Oral Mucosal Epithelial Cells. Front. Immunol. 2019, 10, 208. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.; Tenenbaum, H.C.; Wong, B.K.C.; Schmitt, C.; Nogueira-Filho, G. Is Keratinized Mucosa Indispensable to Maintain Peri-Implant Health? A Systematic Review of the Literature. J. Biomed. Mater. Res. B Appl. Biomater. 2014, 102, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Ono, M.; Hara, E.S.; Komori, T.; Edamatsu, M.; Yonezawa, T.; Kimura-Ono, A.; Maekawa, K.; Kuboki, T.; Oohashi, T. Type XVIII Collagen Modulates Keratohyalin Granule Formation and Keratinization in Oral Mucosa. Int. J. Mol. Sci. 2019, 20, 4739. [Google Scholar] [CrossRef]
- Presland, R.B.; Jurevic, R.J. Making Sense of the Epithelial Barrier: What Molecular Biology and Genetics Tell Us about the Functions of Oral Mucosal and Epidermal Tissues. J. Dent. Educ. 2002, 66, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.; Divo, M.; Langbein, L. The Human Keratins: Biology and Pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef] [PubMed]
- Bragulla, H.H.; Homberger, D.G. Structure and Functions of Keratin Proteins in Simple, Stratified, Keratinized and Cornified Epithelia. J. Anat. 2009, 214, 516–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yin, M.; Zhang, L.J. Keratin 6, 16 and 17—Critical Barrier Alarmin Molecules in Skin Wounds and Psoriasis. Cells 2019, 8, 807. [Google Scholar] [CrossRef]
- Dawson, D.V.; Drake, D.R.; Hill, J.R.; Brogden, K.A.; Fischer, C.L.; Wertz, P.W. Organization, Barrier Function and Antimicrobial Lipids of the Oral Mucosa. Int. J. Cosmet. Sci. 2013, 35, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, K.; Machida, H.; Nakagawa, A.; Ahn, K.; Morita, R.; Sekiguchi, K.; Miner, J.H.; Fujiwara, H. Mapping the Molecular and Structural Specialization of the Skin Basement Membrane for Inter-Tissue Interactions. Nat. Commun. 2021, 12, 2577. [Google Scholar] [CrossRef]
- Pereira, D.; Sequeira, I. A Scarless Healing Tale: Comparing Homeostasis and Wound Healing of Oral Mucosa with Skin and Oesophagus. Front. Cell Dev. Biol. 2021, 9, 682143. [Google Scholar] [CrossRef]
- Canalis, E.; Economides, A.N.; Gazzerro, E. Bone Morphogenetic Proteins, Their Antagonists, and the Skeleton. Endocr. Rev. 2003, 24, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Chung, C.H.; Lu, Y.C.; Wu, M.H.; Chou, P.H.; Yen, J.Y.; Lai, Y.W.; Wang, G.S.; Liu, S.C.; Cheng, J.K.; et al. BMP-2 Induces Angiogenesis by Provoking Integrin A6 Expression in Human Endothelial Progenitor Cells. Biochem. Pharmacol. 2018, 150, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Gosselet, F.P.; Magnaldo, T.; Culerrier, R.M.; Sarasin, A.; Ehrhart, J.C. BMP2 and BMP6 Control P57Kip2 Expression and Cell Growth Arrest/Terminal Differentiation in Normal Primary Human Epidermal Keratinocytes. Cell Signal 2007, 19, 731–739. [Google Scholar] [CrossRef]
- Itoh, K.; Kataoka, H.; Sasaki, M.; Tanida, S.; Oshima, T.; Ogasawara, N.; Ohara, H.; Nakao, H.; Joh, T. Bone Morphogenetic Protein 2 Induced Differentiation toward Superficial Epithelial Cells in the Gastric Mucosa. J. Gastroenterol. 2006, 41, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Tardaguila, M.; De La Fuente, L.; Marti, C.; Pereira, C.; Pardo-Palacios, F.J.; Del Risco, H.; Ferrell, M.; Mellado, M.; Macchietto, M.; Verheggen, K.; et al. SQANTI: Extensive Characterization of Long-Read Transcript Sequences for Quality Control in Full-Length Transcriptome Identification and Quantification. Genome Res. 2018, 28, 1096. [Google Scholar] [CrossRef] [PubMed]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved Transcriptome Assembly Using a Hybrid of Long and Short Reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A.; Berger, B. IsoformSwitchAnalyzeR: Analysis of Changes in Genome-Wide Patterns of Alternative Splicing and Its Functional Consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Basso, F.G.; Soares, D.G.; Pansani, T.N.; Turrioni, A.P.S.; Scheffel, D.L.; Hebling, J.; Costa, C.A.D.S. Response of a Co-Culture Model of Epithelial Cells and Gingival Fibroblasts to Zoledronic Acid. Braz. Oral. Res. 2016, 30, e122. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Ono, M.; Hara, E.S.; Ueda, J.; Nguyen, H.T.T.; Nguyen, H.T.; Yonezawa, T.; Maeba, T.; Kimura-Ono, A.; Takarada, T.; et al. Type IV Collagen A6 Chain Is a Regulator of Keratin 10 in Keratinization of Oral Mucosal Epithelium. Sci. Rep. 2018, 8, 2612. [Google Scholar] [CrossRef] [PubMed]
- Lowery, J.W.; Rosen, V. The BMP Pathway and Its Inhibitors in the Skeleton. Physiol. Rev. 2018, 98, 2431–2452. [Google Scholar] [CrossRef]
- Mukherjee, S.; Katarkar, A.; Dhariwal, R.; Mohanty, S.; Mahato, B.; Ray, J.G.; Chaudhuri, K. Effect of Lysyl Oxidase G473A Polymorphism on Lysyl Oxidase and Total Soluble Collagen Expression in Oral Submucous Fibrosis. Asian Pac. J. Cancer Prev. 2021, 22, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Shen, W.; Shi, X.; Wang, Q.; Zhu, L.; Xu, X.; Yu, J.; Liu, L. Upregulated LOX and Increased Collagen Content Associated with Aggressive Clinicopathological Features and Unfavorable Outcome in Oral Squamous Cell Carcinoma. J. Cell Biochem. 2019, 120, 14348–14359. [Google Scholar] [CrossRef] [PubMed]
- Le Provost, G.S.; Debret, R.; Cenizo, V.; Aimond, G.; Pez, F.; Kaniewski, B.; André, V.; Sommer, P. Lysyl Oxidase Silencing Impairs Keratinocyte Differentiation in a Reconstructed-Epidermis Model. Exp. Dermatol. 2010, 19, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.S.; Yang, J.R.; Liao, S.C.; Hsu, C.C.; Hsu, C.W.; Yuan, K. Latent Transforming Growth Factor-β Binding Proteins (LTBP-1 and LTBP-2) and Gingiva Keratinization. Oral. Dis. 2015, 21, 762–769. [Google Scholar] [CrossRef]
- Przyklenk, M.; Georgieva, V.S.; Metzen, F.; Mostert, S.; Kobbe, B.; Callewaert, B.; Sengle, G.; Brachvogel, B.; Mecham, R.P.; Paulsson, M.; et al. LTBP1 Promotes Fibrillin Incorporation into the Extracellular Matrix. Matrix Biol. 2022, 110, 60–75. [Google Scholar] [CrossRef]
- Wang, Y.; Tatakis, D.N. Human Gingiva Transcriptome during Wound Healing. J. Clin. Periodontol. 2017, 44, 394–402. [Google Scholar] [CrossRef]
- Rubina, K.A.; Sysoeva, V.Y.; Zagorujko, E.I.; Tsokolaeva, Z.I.; Kurdina, M.I.; Parfyonova, Y.V.; Tkachuk, V.A. Increased Expression of UPA, UPAR, and PAI-1 in Psoriatic Skin and in Basal Cell Carcinomas. Arch. Dermatol. Res. 2017, 309, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, Z. MicroRNA 617 Targeting SERPINE1 Inhibited the Progression of Oral Squamous Cell Carcinoma. Mol. Cell Biol. 2021, 41, e00565-20. [Google Scholar] [CrossRef] [PubMed]
- Hamajima, Y.; Komori, M.; Preciado, D.A.; Choo, D.I.; Moribe, K.; Murakami, S.; Ondrey, F.G.; Lin, J. The Role of Inhibitor of DNA-Binding (Id1) in Hyperproliferation of Keratinocytes: The Pathological Basis for Middle Ear Cholesteatoma from Chronic Otitis Media. Cell Prolif. 2010, 43, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Mark, E.B.; Jonsson, M.; Asp, J.; Wennberg, A.M.; Mölne, L.; Lindahl, A. Expression of Genes Involved in the Regulation of P16 in Psoriatic Involved Skin. Arch. Dermatol. Res. 2006, 297, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Varani, J.; Perone, P.; Deming, M.O.B.; Warner, R.L.; Aslam, M.N.; Bhagavathula, N.; Dame, M.K.; Voorhees, J.J. Impaired Keratinocyte Function on Matrix Metalloproteinase-1 (MMP-1) Damaged Collagen. Arch. Dermatol. Res. 2009, 301, 497–506. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Kwon, K.S.; Moon, H.E.; Park, J.A.; Choi, K.S.; Kim, Y.S.; Jang, H.S.; Oh, C.K.; Lee, Y.M.; Kwon, Y.G.; et al. Insulin-like Growth Factor-II Regulates the Expression of Vascular Endothelial Growth Factor by the Human Keratinocyte Cell Line HaCaT. J. Investig. Dermatol. 2004, 123, 152–158. [Google Scholar] [CrossRef]
- Cacheux, W.; Lièvre, A.; Richon, S.; Vacher, S.; El Alam, E.; Briaux, A.; El Botty, R.; Mariani, P.; Buecher, B.; Schnitzler, A.; et al. Interaction between IGF2-PI3K Axis and Cancer-Associated-Fibroblasts Promotes Anal Squamous Carcinogenesis. Int. J. Cancer 2019, 145, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, T.; Wang, F.; Gong, B.; Xie, W.; Ma, M.; Yang, X. Long Noncoding RNA IGF2AS Is Acting as an Epigenetic Tumor Suppressor in Human Prostate Cancer. Urology 2019, 124, 310.e1–310.e8. [Google Scholar] [CrossRef]
- He, M.; Liang, P. IL-24 Transgenic Mice: In Vivo Evidence of Overlapping Functions for IL-20, IL-22, and IL-24 in the Epidermis. J. Immunol. 2010, 184, 1793–1798. [Google Scholar] [CrossRef] [PubMed]
- Mitamura, Y.; Nunomura, S.; Nanri, Y.; Ogawa, M.; Yoshihara, T.; Masuoka, M.; Tsuji, G.; Nakahara, T.; Hashimoto-Hachiya, A.; Conway, S.J.; et al. The IL-13/Periostin/IL-24 Pathway Causes Epidermal Barrier Dysfunction in Allergic Skin Inflammation. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Bosanquet, D.C.; Harding, K.G.; Ruge, F.; Sanders, A.J.; Jiang, W.G. Expression of IL-24 and IL-24 Receptors in Human Wound Tissues and the Biological Implications of IL-24 on Keratinocytes. Wound Repair. Regen. 2012, 20, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Adzraku, S.Y.; Wang, G.; Cao, C.; Bao, Y.; Wang, Y.; Smith, A.O.; Du, Y.; Wang, H.; Li, Y.; Xu, K.; et al. Robo4 Inhibits Gamma Radiation-Induced Permeability of a Murine Microvascular Endothelial Cell by Regulating the Junctions. Cell Mol. Biol. Lett. 2023, 28, 2. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.P.; You, J.S.; Oh, J.S. Technical Note on Simplified Free Gingival Graft Using Tack Fixation (SFGG). Medicina 2023, 59, 2062. [Google Scholar] [CrossRef] [PubMed]
- Glim, J.E.; Everts, V.; Niessen, F.B.; Ulrich, M.M.; Beelen, R.H.J. Extracellular Matrix Components of Oral Mucosa Differ from Skin and Resemble That of Foetal Skin. Arch. Oral. Biol. 2014, 59, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Sabapaty, A.; Lin, P.Y.; Dunn, J.C.Y. Effect of Air–Liquid Interface on Cultured Human Intestinal Epithelial Cells. FASEB Bioadv. 2024, 6, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Caddy, J.C.; Luoma, L.M.; Berry, F.B. FOXC1 Negatively Regulates BMP-SMAD Activity and Id1 Expression during Osteoblast Differentiation. J. Cell Biochem. 2020, 121, 3266–3277. [Google Scholar] [CrossRef]
- Salazar, V.S.; Zarkadis, N.; Huang, L.; Norris, J.; Grimston, S.K.; Mbalaviele, G.; Civitelli, R. Embryonic Ablation of Osteoblast Smad4 Interrupts Matrix Synthesis in Response to Canonical Wnt Signaling and Causes an Osteogenesis-Imperfectalike Phenotype. J. Cell Sci. 2013, 126, 4974–4984. [Google Scholar] [CrossRef]

| Gene Symbol | Cell Type/Disease | Function | Reference |
|---|---|---|---|
| LOX | Oral submucous fibrosis, Oral squamous cell carcinoma | Induce deposition of collagen component. | [26,27] |
| Epidermal keratinocytes | Induce epithelial differentiation. | [28] | |
| LTBP1 | Oral mucosal epithelial cells | Induce epithelial differentiation. | [29] |
| Cutaneous fibroblasts | Promotes fibrillin incorporation into the extracellular matrix. | [30] | |
| SERPINE1 | Gingival wound healing | Upregulated during wound healing. | [31] |
| Psoriatic skin, basal cell carcinoma | Upregulated in lesions (hyperproliferation) than normal skin. | [32] | |
| Oral squamous cell carcinoma | Enhance cell proliferation; Inhibit cell apoptosis. | [33] | |
| ID1 | Epidermal keratinocytes | Induce cell proliferation. | [34] |
| Psoriatic skin | Upregulated in epidermis than normal skin. | [35] | |
| MMP1 | Epidermal keratinocytes | Induce cell differentiation (E-cadherin) via collagen fragmentation. | [36] |
| Gingival wound healing | Upregulated during wound healing. | [31] | |
| IGF2 | Psoriasis | Induce angiogenesis. | [37] |
| Anal squamous cell carcinoma, Prostate cancer | Induce cell proliferation. | [38,39] | |
| IL24 | Epidermal keratinocytes | Induce keratinocyte proliferation and hyperplasia (KRT6, KRT16); inhibit epidermal late differentiation (filaggrin, loricrin). | [40,41] |
| Cutaneous wound healing | Inhibit cell migration. | [42] | |
| ROBO4 | Endothelial cell | Induce junctional adhesion molecules (ZO-1, occludin). | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, X.; Ono, M.; Nguyen, H.T.T.; Wang, Z.; Zhao, K.; Komori, T.; Yonezawa, T.; Kuboki, T.; Oohashi, T. Exploring the Regulators of Keratinization: Role of BMP-2 in Oral Mucosa. Cells 2024, 13, 807. https://doi.org/10.3390/cells13100807
Mu X, Ono M, Nguyen HTT, Wang Z, Zhao K, Komori T, Yonezawa T, Kuboki T, Oohashi T. Exploring the Regulators of Keratinization: Role of BMP-2 in Oral Mucosa. Cells. 2024; 13(10):807. https://doi.org/10.3390/cells13100807
Chicago/Turabian StyleMu, Xindi, Mitsuaki Ono, Ha Thi Thu Nguyen, Ziyi Wang, Kun Zhao, Taishi Komori, Tomoko Yonezawa, Takuo Kuboki, and Toshitaka Oohashi. 2024. "Exploring the Regulators of Keratinization: Role of BMP-2 in Oral Mucosa" Cells 13, no. 10: 807. https://doi.org/10.3390/cells13100807