Thirteen Ovary-Enriched Genes Are Individually Not Essential for Female Fertility in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. In Silico Gene Expression Analysis of Candidate Genes
2.3. Generation of Knockout Mice with the CRISPR/Cas9 System
2.4. Fertility Analysis of KO Lines
2.5. Histological Analysis of Ovaries
2.6. In Vitro Fertilization (IVF)
2.7. Statistical Analyses
3. Results
3.1. Expression Patterns of Candidate Genes in Mice
3.2. Fertility Results of 13 Ovary-Enriched KO Mouse Models
3.3. Phenotypic Analysis of Ccdc58 KO Mouse Line
3.4. Phenotypic Analysis of D930020B18Rik KO Mouse Line
3.5. Phenotypic Analysis of Elobl KO Mouse Line
3.6. Phenotypic Analysis of Oas1h KO Mouse Line
3.7. Phenotypic Analysis of Pkd1l2 KO Mouse Line
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vander Borght, M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef]
- World Health Organization. Infertility Prevalence Estimates: 1990–2021; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- Walker, M.H.; Tobler, K.J. Female Infertility. In StatPearls [Internet]; Updated 19 December 2022; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Recent Advances in Medically Assisted Conception; Report of a WHO Scientific Group; World Health Organization Technical Report Series; WHO: Geneva, Switzerland, 1992; Volume 820, pp. 1–111.
- Quaas, A.; Dokras, A. Diagnosis and treatment of unexplained infertility. Rev. Obstet. Gynecol. 2008, 1, 69–76. [Google Scholar]
- Zorrilla, M.; Yatsenko, A.N. The Genetics of Infertility: Current Status of the Field. Curr. Genet. Med. Rep. 2013, 1, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Emori, C.; Kodani, M.; Abbasi, F.; Mori, M.; Ikawa, M. PABPN1L is required for maternal mRNA degradation after meiosis resumption. J. Reprod. Dev. 2023, 70, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhu, Y.; Chen, H.; Wu, Y.; Pi, S.; Chen, L.; Shen, L.; Fan, H. PABPN1L mediates cytoplasmic mRNA decay as a placeholder during the maternal-to-zygotic transition. Embo Rep. 2020, 21, e49956. [Google Scholar] [CrossRef]
- De Baere, E.; Beysen, D.; Oley, C.; Lorenz, B.; Cocquet, J.; De Sutter, P.; Devriendt, K.; Dixon, M.; Fellous, M.; Fryns, J.-P.; et al. FOXL2 and BPES: Mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am. J. Hum. Genet. 2003, 72, 478–487. [Google Scholar] [CrossRef]
- Abbasi, F.; Kodani, M.; Emori, C.; Kiyozumi, D.; Mori, M.; Fujihara, Y.; Ikawa, M. CRISPR/Cas9-Mediated Genome Editing Reveals Oosp Family Genes are Dispensable for Female Fertility in Mice. Cells 2020, 9, 821. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sáez, F.; Sainz-Urruela, R.; Felipe-Medina, N.; Condezo, Y.B.; Sánchez-Martín, M.; Llano, E.; Pendás, A.M. The Oocyte-Specific Linker Histone H1FOO Is Not Essential for Mouse Oogenesis and Fertility. Cells 2022, 11, 3706. [Google Scholar] [CrossRef]
- Miyata, H.; Castaneda, J.M.; Fujihara, Y.; Yu, Z.; Archambeault, D.R.; Isotani, A.; Kiyozumi, D.; Kriseman, M.L.; Mashiko, D.; Matsumura, T.; et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci. USA 2016, 113, 7704–7710. [Google Scholar] [CrossRef]
- Lu, Y.; Oura, S.; Matsumura, T.; Oji, A.; Sakurai, N.; Fujihara, Y.; Shimada, K.; Miyata, H.; Tobita, T.; Noda, T.; et al. CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice. Biol. Reprod. 2019, 101, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Takeo, T.; Nakagata, N. Superovulation using the combined administration of inhibin antiserum and equine chorionic gonadotropin increases the number of ovulated oocytes in C57BL/6 female mice. PLoS ONE 2015, 10, e0128330. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Wigglesworth, K.; Eppig, J.J.; Schultz, R.M. Preimplantation development of mouse embryos in KSOM: Augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 1995, 41, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Yokoyama, M.; Hosi, T. Studies on the fertilization of mouse eggs in vitro. II. Effects of in vitro preincubation of spermatozoa on time of sperm penetration of mouse eggs in vitro. Jpn. J. Anim. Reprod. 1971, 16, 152–157. [Google Scholar] [CrossRef]
- Mahadevan, S.; Sathappan, V.; Utama, B.; Lorenzo, I.; Kaskar, K.; Veyver, I.B.V.D. Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Sci. Rep. 2017, 7, srep44667. [Google Scholar] [CrossRef] [PubMed]
- Kuchmiy, A.A.; D’hont, J.; Hochepied, T.; Lamkanfi, M. NLRP2 controls age-associated maternal fertility. J. Exp. Med. 2016, 213, 2851–2860. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Ma, L.; Stein, P.; Pangas, S.A.; Burns, K.H.; Bai, Y.; Schultz, R.M.; Matzuk, M.M. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol. Cell. Biol. 2005, 25, 4615–4624. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, S.; Shibata, S.; Iwakura, Y. Genomic structure of the mouse 2’,5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2022, 22, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Oura, S.; Mori, H.; Ikawa, M. Genome editing in mice and its application to the study of spermatogenesis. Gene Genome Ed. 2022, 3–4, 100014. [Google Scholar] [CrossRef]
- Zöller, E.; Laborenz, J.; Krämer, L.; Boos, F.; Räschle, M.; Alexander, R.T.; Herrmann, J.M. The intermembrane space protein Mix23 is a novel stress-induced mitochondrial import factor. J. Biol. Chem. 2020, 295, 14686–14697. [Google Scholar] [CrossRef]
- Liu, X.; Salokas, K.; Tamene, F.; Jiu, Y.; Weldatsadik, R.G.; Öhman, T.; Varjosalo, M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Kirillova, A.; Smitz, J.E.J.; Sukhikh, G.T.; Mazunin, I. The Role of Mitochondria in Oocyte Maturation. Cells 2021, 10, 2484. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.; Horvat, F.; Drutovic, D.; Efenberkova, M.; Pinkas, D.; Jindrova, A.; Pasulka, J.; Iyyappan, R.; Malik, R.; Susor, A.; et al. The most abundant maternal lncRNA Sirena1 acts post-transcriptionally and impacts mitochondrial distribution. Nucleic Acids Res. 2020, 48, 3211–3227. [Google Scholar] [CrossRef] [PubMed]
- Dalton, C.M.; Szabadkai, G.; Carroll, J. Measurement of ATP in single oocytes: Impact of maturation and cumulus cells on levels and consumption. J. Cell. Physiol. 2014, 229, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, K.; Montell, C. TRP channels. Annu. Rev. Biochem. 2007, 76, 387–417. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Tian, X.; Sung, S.-W.; Somlo, S. Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics 2003, 81, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, T.; Takakura, A.; Denker, B.M.; Venugopal, B.; Zhou, J. Polycystin-1L2 is a novel G-protein-binding protein. Genomics 2004, 84, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Kokosar, M.; Benrick, A.; Perfilyev, A.; Fornes, R.; Nilsson, E.; Maliqueo, M.; Behre, C.J.; Sazonova, A.; Ohlsson, C.; Ling, C.; et al. Epigenetic and Transcriptional Alterations in Human Adipose Tissue of Polycystic Ovary Syndrome. Sci. Rep. 2016, 6, 22883. [Google Scholar] [CrossRef]
- Peng, H.; Chang, B.; Lu, C.; Su, J.; Wu, Y.; Lv, P.; Wang, Y.; Liu, J.; Zhang, B.; Quan, F.; et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS ONE 2012, 7, e30344. [Google Scholar] [CrossRef]
- Rong, Y.; Ji, S.-Y.; Zhu, Y.-Z.; Wu, Y.-W.; Shen, L.; Fan, H.-Y. ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res. 2019, 47, 11387–11402. [Google Scholar] [CrossRef] [PubMed]
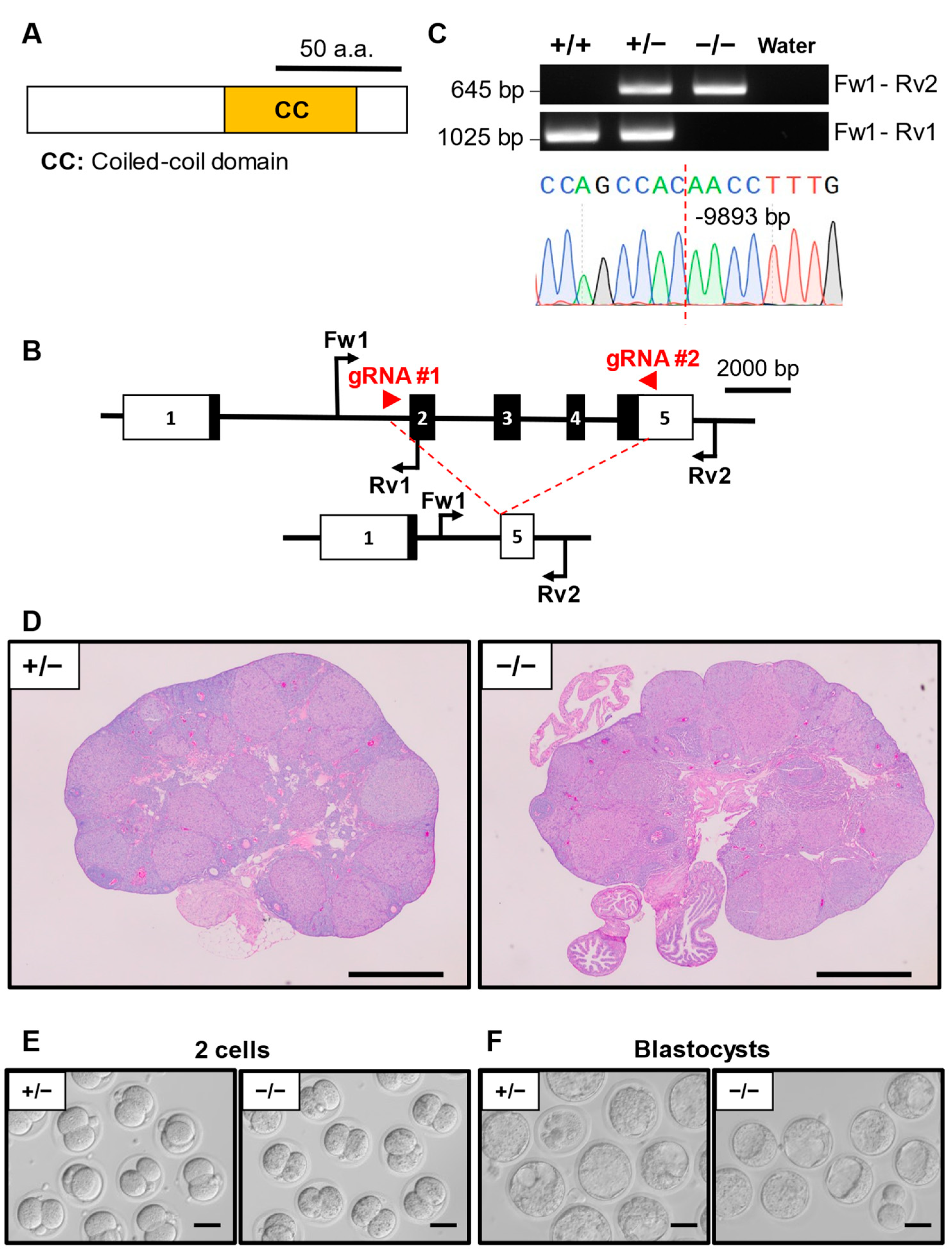

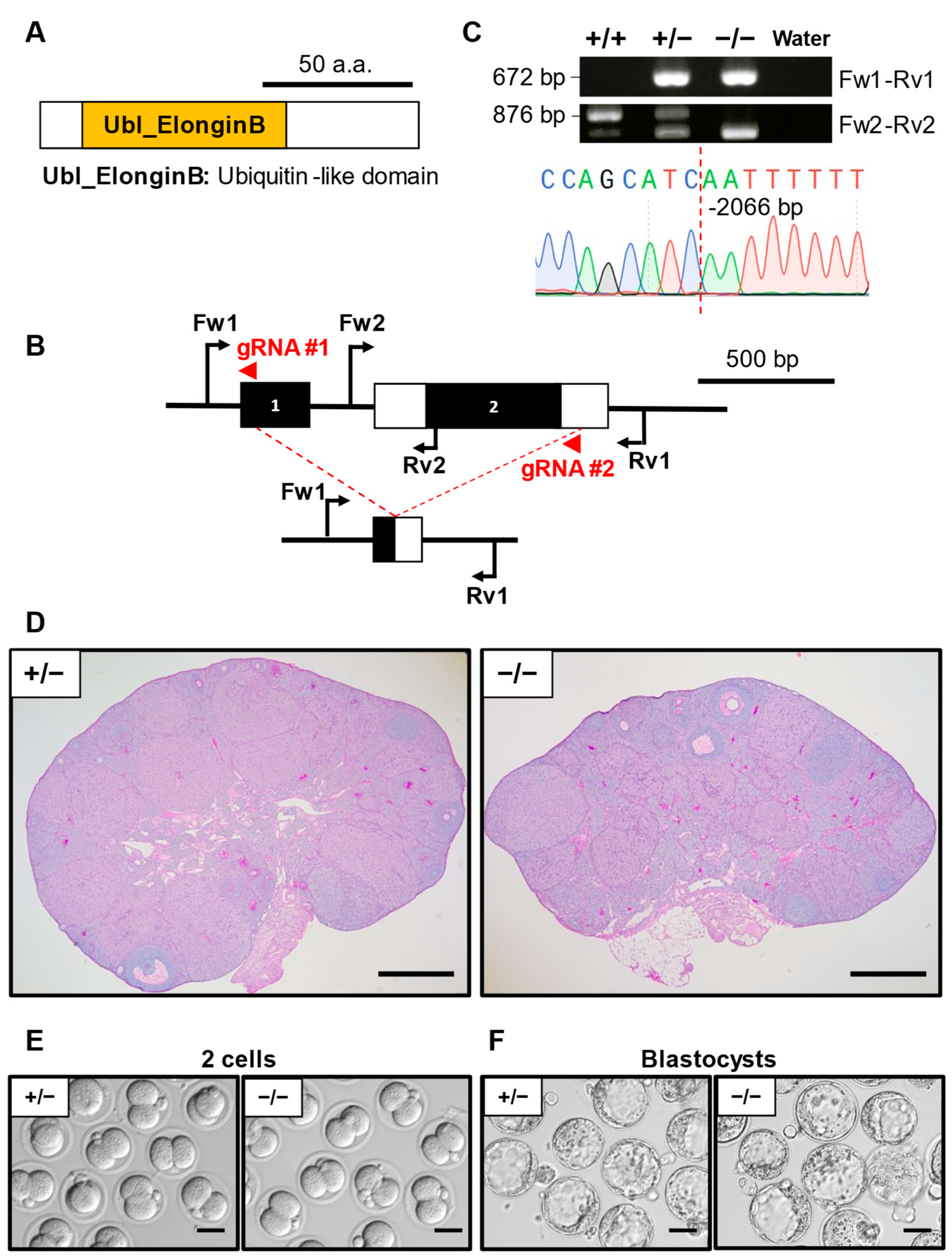
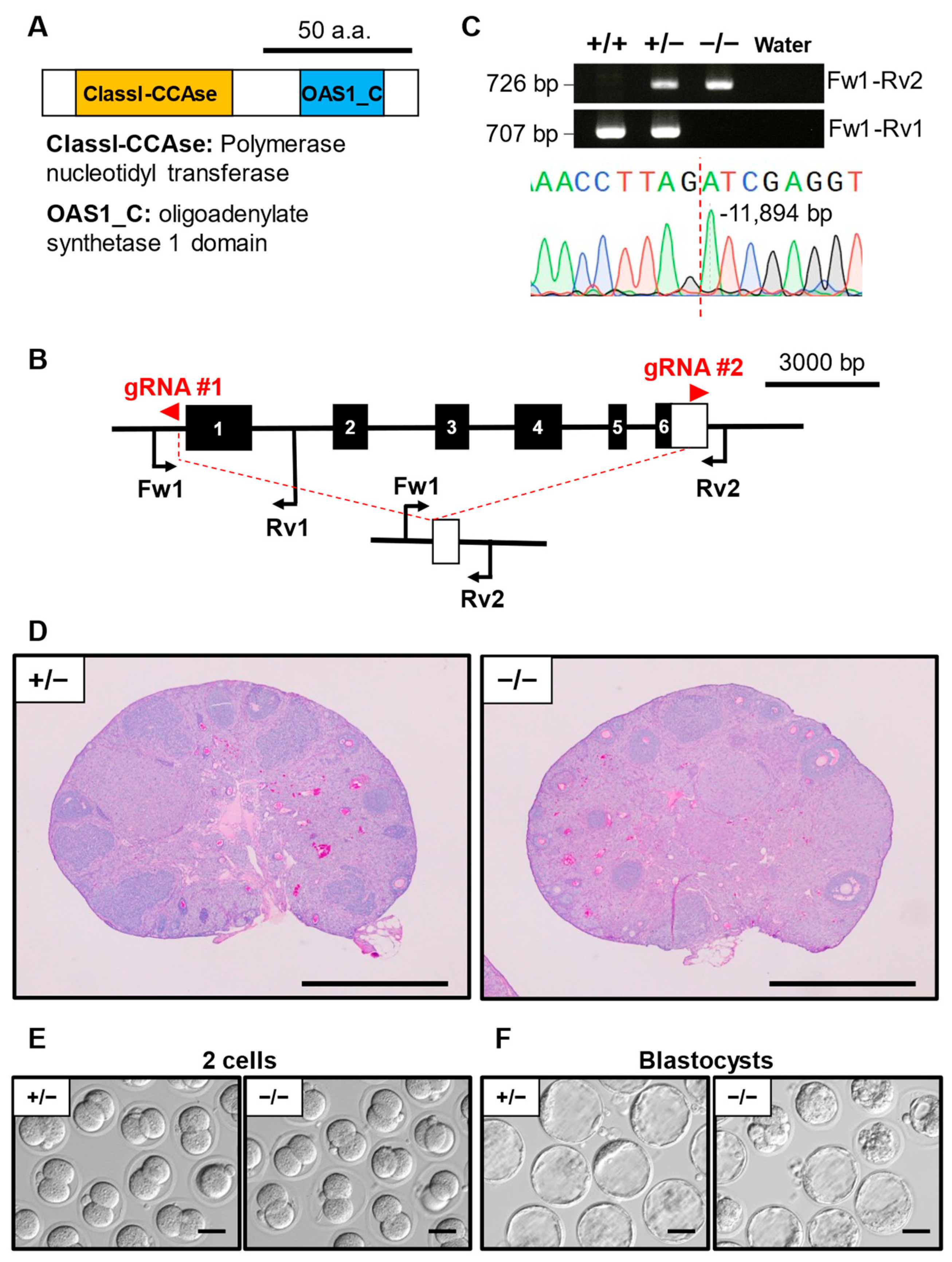
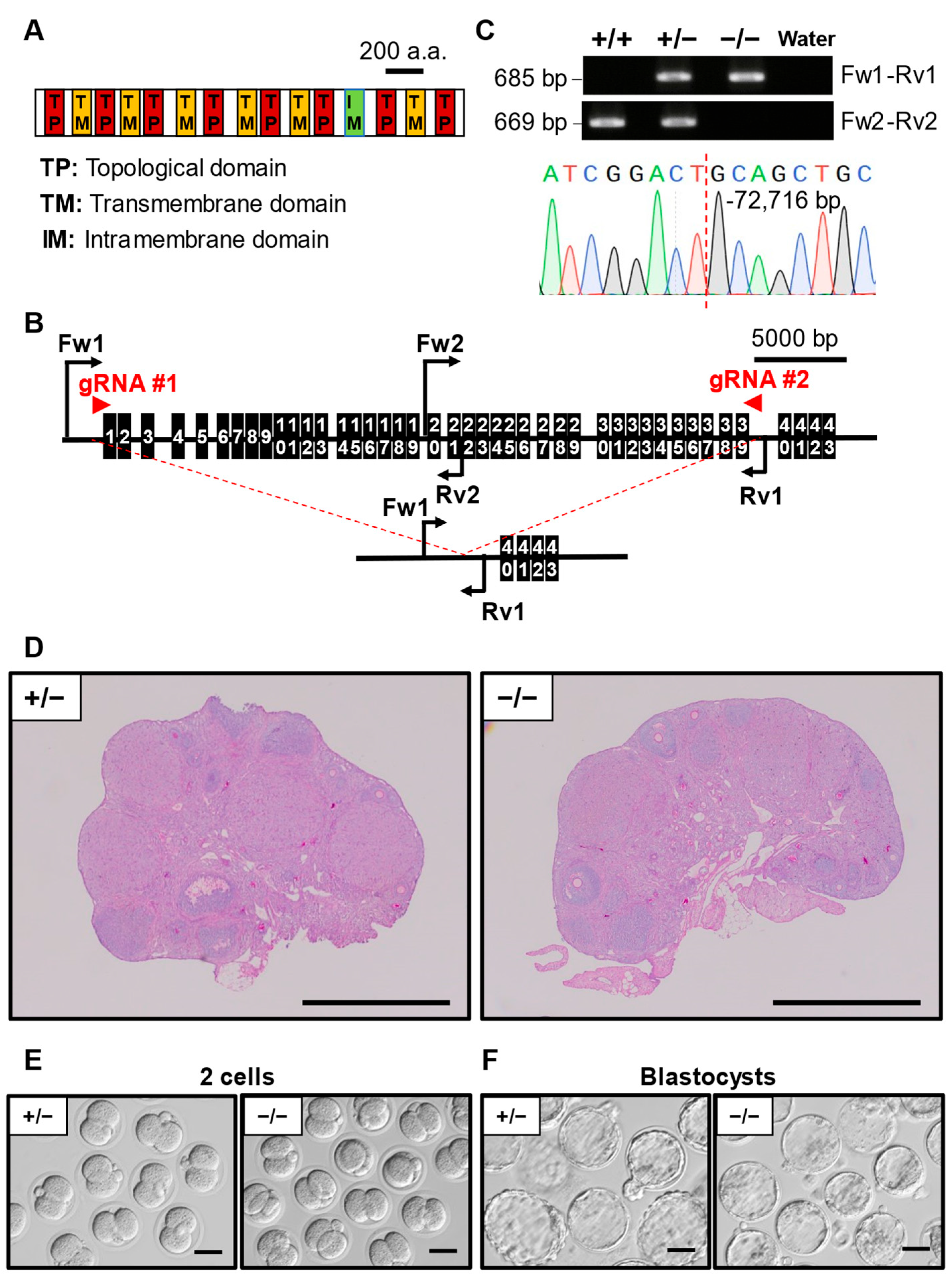
| Gene Symbol | Genotype | No. of Females | No. of Pups | No. of Litters | Average Litter Size ± SD |
|---|---|---|---|---|---|
| Wildtype | 9 | 247 | 30 | 8.2 ± 2.7 | |
| Ccdc58 | −9893/−9893 | 7 | 182 | 23 | 7.9 ± 3.1 |
| D930020B18Rik | −50,969/−50,969 | 5 | 107 | 15 | 7.1 ± 2.4 |
| Elobl | −2066/−2066 | 4 | 134 | 16 | 8.4 ± 3.0 |
| Fbxw15 | −15,610/−15,610 | 3 | 84 | 9 | 9.3 ± 1.1 |
| Nlrp2 | −9171/−9171 | 5 | 81 | 15 | 5.4 ± 2.6 * |
| Oas1h | −11,894/−11,894 | 3 | 85 | 10 | 8.5 ± 2.9 |
| Pkd1l2 | −72,716/−72,716 | 3 | 61 | 9 | 6.8 ± 1.9 |
| Pramel34 | −3037 + 20ins/−3037 + 20ins | 3 | 131 | 16 | 8.2 ± 1.6 |
| Pramel47 | −3616/−3616 | 5 | 198 | 25 | 7.9 ± 2.2 |
| Sting1 | −5916/−5916 | 5 | 98 | 15 | 6.5 ± 2.9 |
| Tspan4 | −10,374/−10,374 | 4 | 119 | 17 | 7.0 ± 3.4 |
| Tubal3 | −3947/−3947 | 6 | 183 | 24 | 7.6 ± 1.8 |
| Zar1l | −11,036/−11,036 | 3 | 77 | 11 | 7.0 ± 2.0 |
| Gene symbol | 2 cell-rate (%) ± SD, N = number of mice, n = number of total oocytes | |
| Het | Hom | |
| Ccdc58 | 89.9 ± 10.0, N = 5, n = 69 | 81.2 ± 20.9, N = 6, n = 62 |
| D930020B18Rik | 88.4 ± 8.6, N = 6, n = 136 | 86.1 ± 12.5, N = 5, n = 109 |
| Elobl | 86.3 ± 13.7, N = 6, n = 106 | 88.8 ± 8.2, N = 7, n = 171 |
| Oas1h | 92.6 ± 12.8, N = 3, n = 49 | 92.2 ± 10.6, N = 5, n = 125 |
| Pkd1l2 | 93.3 ± 4.2, N = 6, n = 97 | 92.2 ± 5.9, N = 7, n = 147 |
| Pramel34 | 82.5 ± 27.7, N = 4, n = 157 | 91.9 ± 4.9, N = 5, n = 187 |
| Pramel47 | 89.2 ± 3.3, N = 4, n = 56 | 81.2 ± 14.5, N = 7, n = 116 |
| Tubal3 | 80.3 ± 17.2, N = 5, n = 101 | 87.0 ± 10.8, N = 7, n = 162 |
| Gene symbol | Blastocyst rate (%) ± SD, N = number of mice, n = number of total oocytes | |
| Het | Hom | |
| Ccdc58 | 79.0 ± 15.9, N = 5, n = 69 | 64.7 ± 16.0, N = 6, n = 62 |
| D930020B18Rik | 81.9 ± 6.3, N = 6, n = 136 | 83.1 ± 9.9, N = 5, n = 109 |
| Elobl | 80.1 ± 19.4, N = 6, n = 106 | 78.8 ± 12.2, N = 7, n = 171 |
| Oas1h | 92.6 ± 12.8, N = 3, n = 49 | 66.6 ± 15.0 *, N = 5, n = 125 |
| Pkd1l2 | 90.0 ± 7.0, N = 6, n = 97 | 85.8 ± 7.6, N = 7, n = 147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, A.H.; Emori, C.; Ishikawa-Yamauchi, Y.; Tokuhiro, K.; Kamoshita, M.; Fujihara, Y.; Ikawa, M. Thirteen Ovary-Enriched Genes Are Individually Not Essential for Female Fertility in Mice. Cells 2024, 13, 802. https://doi.org/10.3390/cells13100802
Pham AH, Emori C, Ishikawa-Yamauchi Y, Tokuhiro K, Kamoshita M, Fujihara Y, Ikawa M. Thirteen Ovary-Enriched Genes Are Individually Not Essential for Female Fertility in Mice. Cells. 2024; 13(10):802. https://doi.org/10.3390/cells13100802
Chicago/Turabian StylePham, Anh Hoang, Chihiro Emori, Yu Ishikawa-Yamauchi, Keizo Tokuhiro, Maki Kamoshita, Yoshitaka Fujihara, and Masahito Ikawa. 2024. "Thirteen Ovary-Enriched Genes Are Individually Not Essential for Female Fertility in Mice" Cells 13, no. 10: 802. https://doi.org/10.3390/cells13100802





