Clorfl86/RHEX Is a Negative Regulator of SCF/KIT Signaling in Human Skin Mast Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Treatments
2.2. Accell®-Mediated RNA Interference
2.3. Immunoblot Analysis
2.4. YoPro-1/Propidium Iodide (PI) Staining
2.5. Caspase-3 Activity
2.6. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.7. Statistics
3. Results
3.1. RHEX Is Highly Expressed in MCs
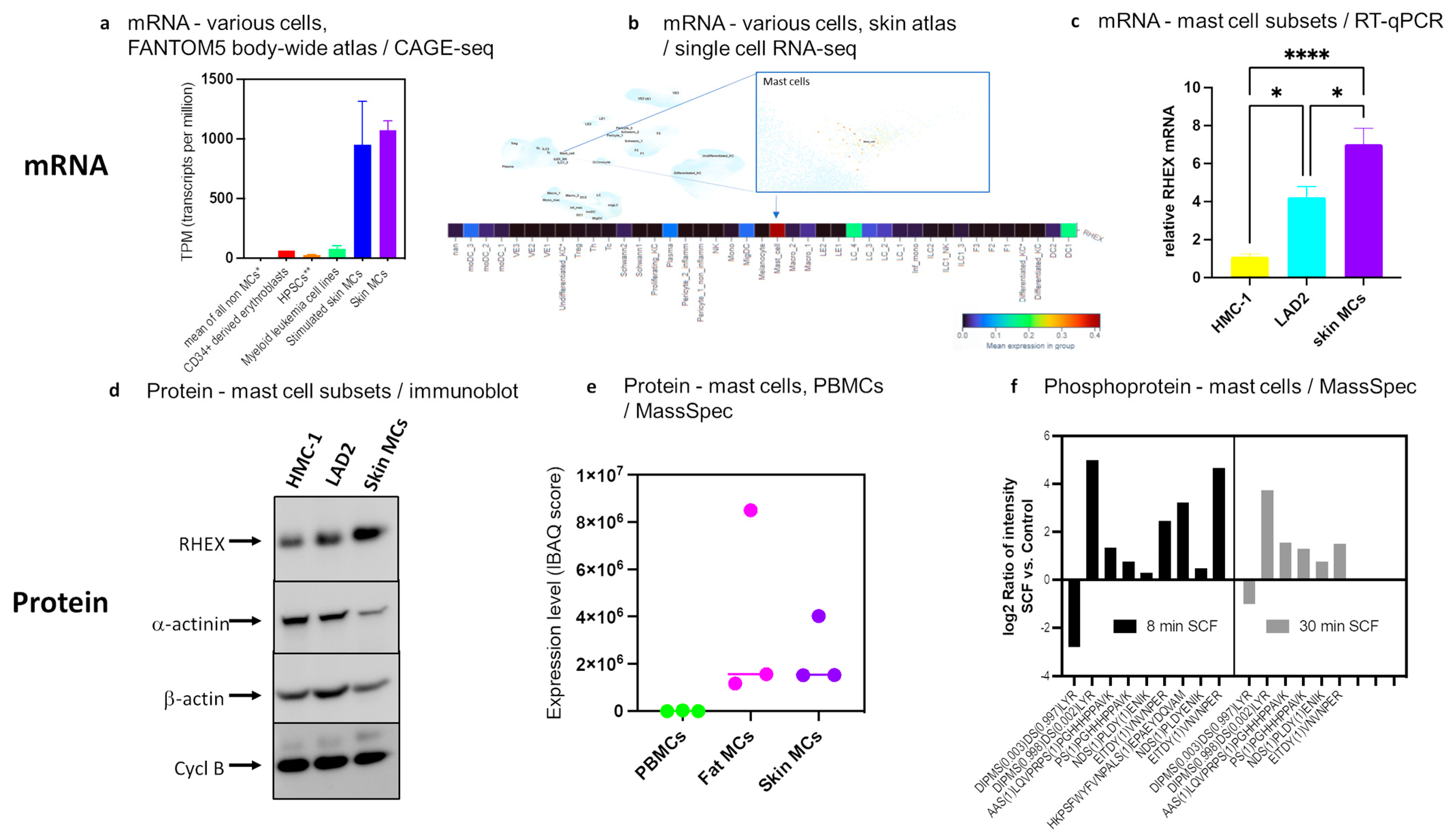
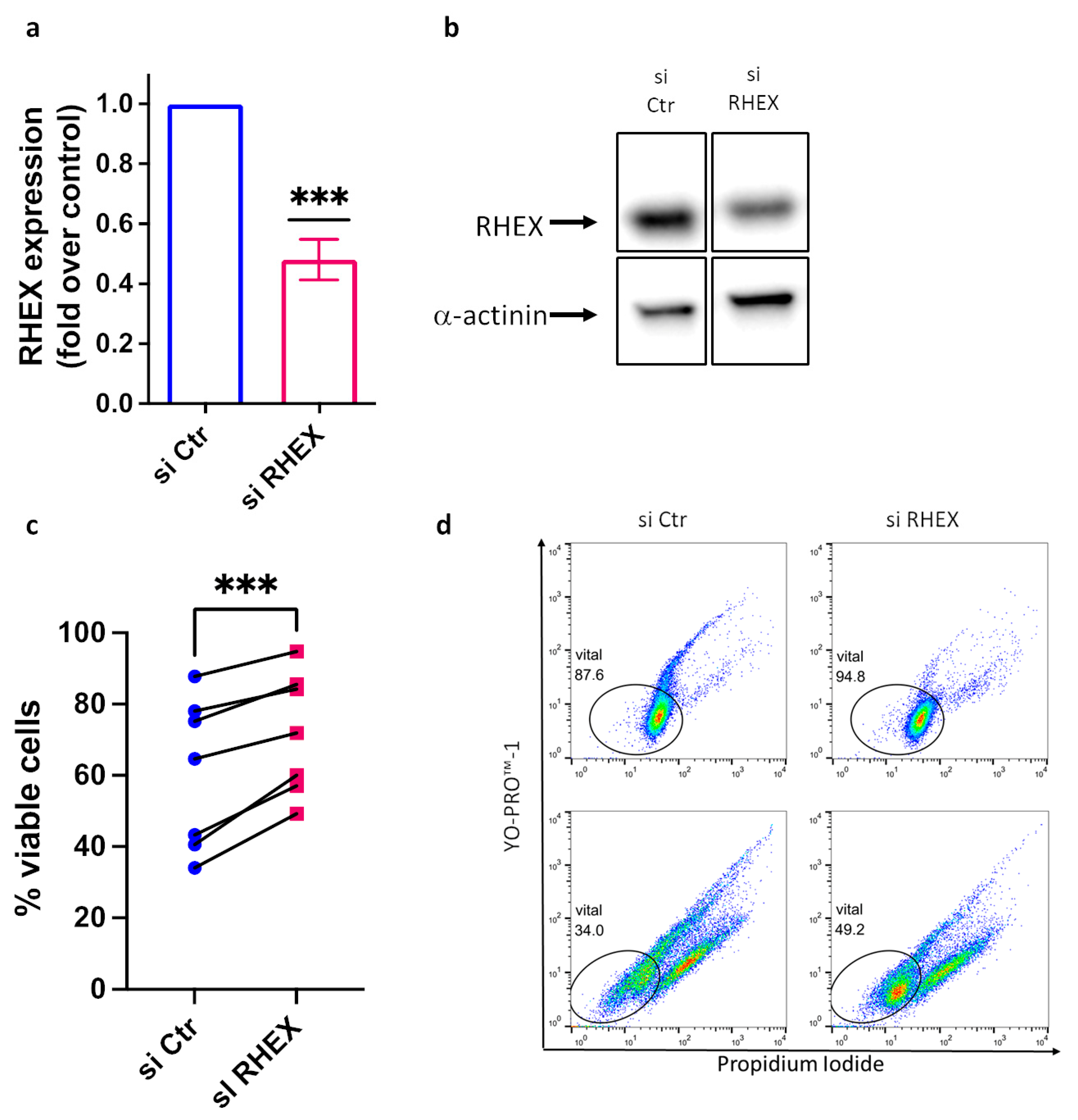
3.2. RHEX Attenuation Supports MC Survival
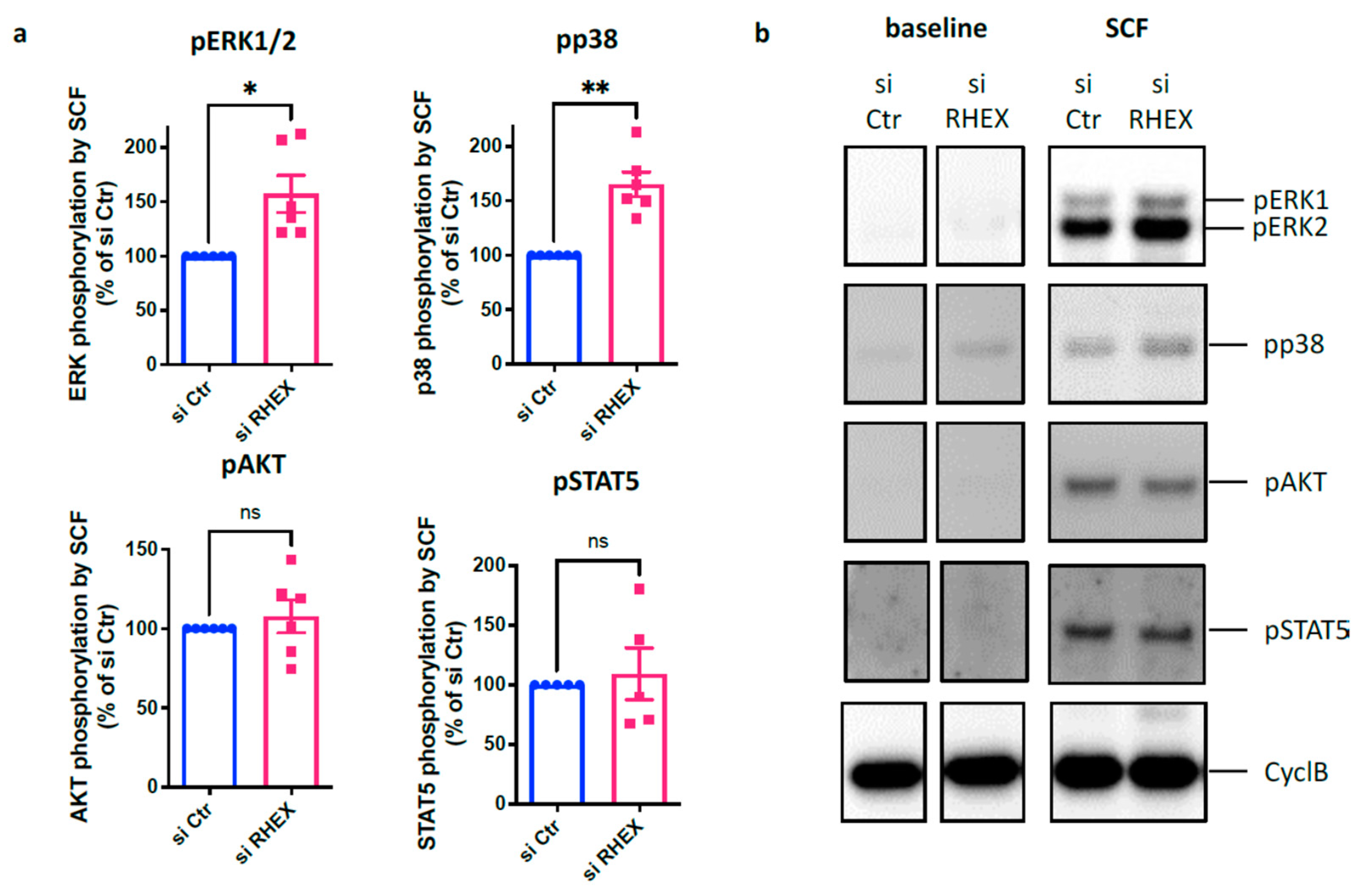
3.3. RHEX Is a Negative Regulator of KIT Signaling Restricting Activation of p38 and ERK, but Not of STAT5 or AKT
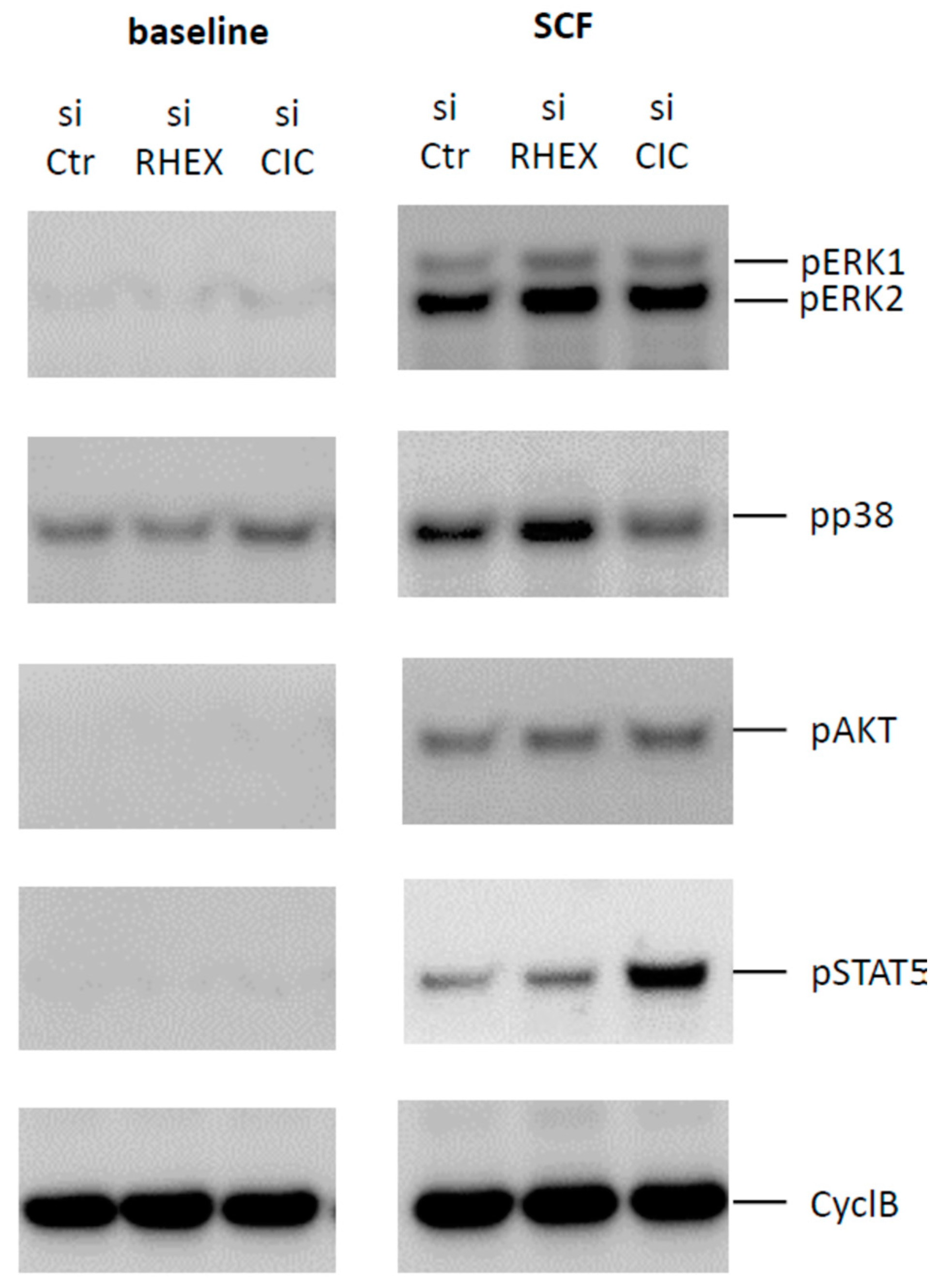
3.4. Comparison between Capicua (CIC) and RHEX: Similar Inhibition of ERK by Both Proteins, but Striking Difference at STAT5
3.5. RHEX Attenuates SCF-Triggered Expression of Immediate-Early Genes (IEGs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Babina, M.; Franke, K.; Bal, G. How “Neuronal” Are Human Skin Mast Cells? Int. J. Mol. Sci. 2022, 23, 10871. [Google Scholar] [CrossRef] [PubMed]
- Gilfillan, A.M.; Beaven, M.A. Regulation of Mast Cell Responses in Health and Disease. Crit. Rev. Immunol. 2011, 31, 475–530. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Cato, A.C.; Ainooson, G.K.; Freichel, M.; Tsvilovskyy, V.; Jessberger, R.; Riedlinger, E.; Sommerhoff, C.P.; Bischoff, S.C. Regulation of the pleiotropic effects of tissue-resident mast cells. J. Allergy Clin. Immunol. 2019, 144, S31–S45. [Google Scholar] [CrossRef] [PubMed]
- Minai-Fleminger, Y.; Levi-Schaffer, F. Mast cells and eosinophils: The two key effector cells in allergic inflammation. Inflamm. Res. 2009, 58, 631–638. [Google Scholar] [CrossRef]
- Metcalfe, D.D.; Peavy, R.D.; Gilfillan, A.M. Mechanisms of mast cell signaling in anaphylaxis. J. Allergy Clin. Immunol. 2009, 124, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 2012, 18, 693–704. [Google Scholar] [CrossRef]
- Kühn, H.; Kolkhir, P.; Babina, M.; Düll, M.; Frischbutter, S.; Fok, J.S.; Jiao, Q.; Metz, M.; Scheffel, J.; Wolf, K.; et al. Mas-related G protein–coupled receptor X2 and its activators in dermatologic allergies. J. Allergy Clin. Immunol. 2020, 147, 456–469. [Google Scholar] [CrossRef]
- Babina, M. The pseudo-allergic/neurogenic route of mast cell activation via MRGPRX2: Discovery, functional programs, regulation, relevance to disease, and relation with allergic stimulation. Itch 2020, 5, e32. [Google Scholar] [CrossRef]
- Roy, S.; Na Ayudhya, C.C.; Thapaliya, M.; Deepak, V.; Ali, H. Multifaceted MRGPRX2: New insight into the role of mast cells in health and disease. J. Allergy Clin. Immunol. 2021, 148, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Aich, A.; Afrin, L.B.; Gupta, K. Mast Cell-Mediated Mechanisms of Nociception. Int. J. Mol. Sci. 2015, 16, 29069–29092. [Google Scholar] [CrossRef]
- Galli, S.J.; Tsai, M. Mast cells in allergy and infection: Versatile effector and regulatory cells in innate and adaptive immunity. Eur. J. Immunol. 2010, 40, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Olivera, A.; Beaven, M.A.; Metcalfe, D.D. Mast cells signal their importance in health and disease. J. Allergy Clin. Immunol. 2018, 142, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Akin, C. Mast cell activation syndromes. J. Allergy Clin. Immunol. 2017, 140, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Shtessel, M.; Limjunyawong, N.; Oliver, E.T.; Chichester, K.; Gao, L.; Dong, X.; Saini, S.S. MRGPRX2 Activation Causes Increased Skin Reactivity in Patients with Chronic Spontaneous Urticaria. J. Investig. Dermatol. 2020, 141, 678–681.e2. [Google Scholar] [CrossRef] [PubMed]
- Worm, M.; Babina, M.; Hompes, S. Causes and risk factors for anaphylaxis. JDDG: J. der Dtsch. Dermatol. Ges. 2012, 11, 44–50. [Google Scholar] [CrossRef]
- Maurer, M.; Eyerich, K.; Eyerich, S.; Ferrer, M.; Gutermuth, J.; Hartmann, K.; Jakob, T.; Kapp, A.; Kolkhir, P.; Larenas-Linnemann, D.; et al. Urticaria: Collegium Internationale Allergologicum (CIA) Update 2020. Int. Arch. Allergy Immunol. 2020, 181, 321–333. [Google Scholar] [CrossRef]
- Church, M.K.; Kolkhir, P.; Metz, M.; Maurer, M. The role and relevance of mast cells in urticaria. Immunol. Rev. 2018, 282, 232–247. [Google Scholar] [CrossRef]
- Wang, Z.; Babina, M. MRGPRX2 signals its importance in cutaneous mast cell biology: Does MRGPRX2 connect mast cells and atopic dermatitis? Exp. Dermatol. 2020, 29, 1104–1111. [Google Scholar] [CrossRef]
- Guntern, P.; Eggel, A. Past, present, and future of anti-IgE biologics. Allergy 2020, 75, 2491–2502. [Google Scholar] [CrossRef]
- Eyerich, S.; Metz, M.; Bossios, A.; Eyerich, K. New biological treatments for asthma and skin allergies. Allergy 2019, 75, 546–560. [Google Scholar] [CrossRef]
- Blank, U.; Huang, H.; Kawakami, T. The high affinity IgE receptor: A signaling update. Curr. Opin. Immunol. 2021, 72, 51–58. [Google Scholar] [CrossRef]
- Bulfone-Paus, S.; Nilsson, G.; Draber, P.; Blank, U.; Levi-Schaffer, F. Positive and Negative Signals in Mast Cell Activation. Trends Immunol. 2017, 38, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Riquer, Z.P.; Segura-Villalobos, D.; Ramírez-Moreno, I.G.; Rodríguez, M.J.P.; Lamas, M.; Gonzalez-Espinosa, C. Signal Transduction Pathways Activated by Innate Immunity in Mast Cells: Translating Sensing of Changes into Specific Responses. Cells 2020, 9, 2411. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Metz, M.; Starkl, P.; Marichal, T.; Tsai, M. Mast cells and IgE in defense against lethality of venoms: Possible “benefit” of allergy. Allergo J. Int. 2020, 29, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Okayama, Y.; Kawakami, T. Development, Migration, and Survival of Mast Cells. Immunol. Res. 2006, 34, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Akin, C.; Metcalfe, D.D. The biology of Kit in disease and the application of pharmacogenetics. J. Allergy Clin. Immunol. 2004, 114, 13–19. [Google Scholar] [CrossRef]
- Lennartsson, J.; Rönnstrand, L. Stem Cell Factor Receptor/c-Kit: From Basic Science to Clinical Implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef]
- Cruse, G.; Metcalfe, D.D.; Olivera, A. Functional deregulation of KIT: Link to mast cell proliferative diseases and other neo-plasms. Immunol. Allergy Clin. 2014, 34, 219–237. [Google Scholar]
- Metcalfe, D.D. Mast cells and mastocytosis. Blood 2008, 112, 946–956. [Google Scholar] [CrossRef]
- Franke, K.; Kirchner, M.; Mertins, P.; Zuberbier, T.; Babina, M. The SCF / KIT axis in human mast cells: Capicua acts as potent KIT repressor and ERK predominates PI3K. Allergy 2022, 77, 3337–3349. [Google Scholar] [CrossRef]
- Wilcock, A.; Bahri, R.; Bulfone-Paus, S.; Arkwright, P.D. Mast cell disorders: From infancy to maturity. Allergy 2018, 74, 53–63. [Google Scholar] [CrossRef]
- Verma, R.; Su, S.; McCrann, D.J.; Green, J.M.; Leu, K.; Young, P.R.; Schatz, P.J.; Silva, J.C.; Stokes, M.P.; Wojchowski, D.M. RHEX, a novel regulator of human erythroid progenitor cell expansion and erythroblast development. J. Exp. Med. 2014, 211, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Wang, Z.; Zuberbier, T.; Babina, M. Cytokines Stimulated by IL-33 in Human Skin Mast Cells: Involvement of NF-kappaB and p38 at Distinct Levels and Potent Co-Operation with FcepsilonRI and MRGPRX2. Int. J. Mol. Sci. 2021, 22, 3580. [Google Scholar] [CrossRef] [PubMed]
- Babina, M.; Wang, Z.; Artuc, M.; Guhl, S.; Zuberbier, T. MRGPRX2 is negatively targeted by SCF and IL-4 to diminish pseudo-allergic stimulation of skin mast cells in culture. Exp. Dermatol. 2018, 27, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Babina, M.; Wang, Z.; Franke, K.; Guhl, S.; Artuc, M.; Zuberbier, T. Yin-Yang of IL-33 in Human Skin Mast Cells: Reduced Degranulation, but Augmented Histamine Synthesis through p38 Activation. J. Investig. Dermatol. 2019, 139, 1516–1525.e3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guhl, S.; Franke, K.; Artuc, M.; Zuberbier, T.; Babina, M. IL-33 and MRGPRX2-Triggered Activation of Human Skin Mast Cells-Elimination of Receptor Expression on Chronic Exposure, but Reinforced Degranulation on Acute Priming. Cells 2019, 8, 341. [Google Scholar] [CrossRef]
- Hazzan, T.; Eberle, J.; Worm, M.; Babina, M. Apoptotic resistance of human skin mast cells is mediated by Mcl-1. Cell Death Discov. 2017, 3, 17048. [Google Scholar] [CrossRef]
- Rastogi, S.; Willmes, D.M.; Nassiri, M.; Babina, M.; Worm, M. PGE2 deficiency predisposes to anaphylaxis by causing mast cell hyperresponsiveness. J. Allergy Clin. Immunol. 2020, 146, 1387–1396.e13. [Google Scholar] [CrossRef]
- Babina, M.; Artuc, M.; Guhl, S.; Zuberbier, T. Retinoic Acid Negatively Impacts Proliferation and MCTC Specific Attributes of Human Skin Derived Mast Cells, but Reinforces Allergic Stimulability. Int. J. Mol. Sci. 2017, 18, 525. [Google Scholar] [CrossRef]
- Guhl, S.; Neou, A.; Artuc, M.; Zuberbier, T.; Babina, M. Skin mast cells develop non-synchronized changes in typical lineage characteristics upon culture. Exp. Dermatol. 2014, 23, 933–935. [Google Scholar] [CrossRef]
- Guhl, S.; Artuc, M.; Neou, A.; Babina, M.; Zuberbier, T. Long-term cultured human skin mast cells are suitable for pharmacological studies of anti-allergic drugs due to high responsiveness to FcepsilonRI cross-linking. Biosci. Biotechnol. Biochem. 2011, 75, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, J.; Weiler, D.; Dewald, G.; Gleich, G. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk. Res. 1988, 12, 345–355. [Google Scholar] [CrossRef]
- Kirshenbaum, A.S.; Akin, C.; Wu, Y.; Rottem, M.; Goff, G.P.; Beaven, M.A.; Rao, V.; Metcalfe, D.D. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of Fcepsilon RI or Fcgamma RI. Leuk. Res. 2003, 27, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Hazzan, T.; Guhl, S.; Artuc, M.; Franke, K.; Worm, M.; Zuberbier, T.; Babina, M. An efficient method for gene knock-down by RNA interference in human skin mast cells. Exp. Dermatol. 2017, 26, 1136–1139. [Google Scholar] [CrossRef] [PubMed]
- Hazzan, T.; Eberle, J.; Worm, M.; Babina, M. Thymic Stromal Lymphopoietin Interferes with the Apoptosis of Human Skin Mast Cells by a Dual Strategy Involving STAT5/Mcl-1 and JNK/Bcl-x(L). Cells 2019, 8, 829. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Bal, G.; Franke, K.; Zuberbier, T.; Babina, M. beta-arrestin-1 and beta-arrestin-2 Restrain MRGPRX2-Triggered Degranulation and ERK1/2 Activation in Human Skin Mast Cells. Front. Allergy 2022, 3, 930233. [Google Scholar] [CrossRef]
- Babina, M.; Wang, Z.; Roy, S.; Guhl, S.; Franke, K.; Artuc, M.; Ali, H.; Zuberbier, T. MRGPRX2 Is the Codeine Receptor of Human Skin Mast Cells: Desensitization through beta-Arrestin and Lack of Correlation with the FcepsilonRI Pathway. J. Investig. Dermatol. 2021, 141, 1286–1296.e4. [Google Scholar] [CrossRef]
- Babina, M.; Wang, Z.; Franke, K.; Zuberbier, T. Thymic Stromal Lymphopoietin Promotes MRGPRX2-Triggered Degranulation of Skin Mast Cells in a STAT5-Dependent Manner with Further Support from JNK. Cells 2021, 10, 102. [Google Scholar] [CrossRef]
- Franke, K.; Bal, G.; Li, Z.; Zuberbier, T.; Babina, M. CREB Is Activated by the SCF/KIT Axis in a Partially ERK-Dependent Manner and Orchestrates Survival and the Induction of Immediate Early Genes in Human Skin Mast Cells. Int. J. Mol. Sci. 2023, 24, 4135. [Google Scholar] [CrossRef]
- Guhl, S.; Stefaniak, R.; Strathmann, M.; Babina, M.; Piazena, H.; Henz, B.M.; Zuberbier, T. Bivalent Effect of UV Light on Human Skin Mast Cells—Low-Level Mediator Release at Baseline but Potent Suppression Upon Mast Cell Triggering. J. Investig. Dermatol. 2005, 124, 453–456. [Google Scholar] [CrossRef]
- Consortium, F.; Baillie, J.K.; de Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; Andersson, R.; Mungall, C.J.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar]
- Kawaji, H.; Kasukawa, T.; Forrest, A.; Carninci, P.; Hayashizaki, Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Sci. Data 2017, 4, 170113. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, G.; Vegh, P.; Fletcher, J.; Poyner, E.F.M.; Stephenson, E.; Goh, I.; Botting, R.A.; Huang, N.; Olabi, B.; Dubois, A.; et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 2021, 371, eaba6500. [Google Scholar] [CrossRef] [PubMed]
- Kirshenbaum, A.S.; Goff, J.P.; Semere, T.; Foster, B.; Scott, L.M. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13). Blood 1999, 94, 2333–2342. [Google Scholar] [CrossRef]
- Guhl, S.; Babina, M.; Neou, A.; Zuberbier, T.; Artuc, M. Mast cell lines HMC-1 and LAD2 in comparison with mature human skin mast cells-drastically reduced levels of tryptase and chymase in mast cell lines. Exp. Dermatol. 2010, 19, 845–847. [Google Scholar] [CrossRef]
- Weber, S.; Babina, M.; Krüger-Krasagakes, S.; Grützkau, A.; Henz, B.M. A subclone (5C6) of the human mast cell line HMC-1 represents a more differentiated phenotype than the original cell line. Arch. Dermatol. Res. 1996, 288, 778–782. [Google Scholar] [CrossRef]
- Dwyer, D.F.; Ordovas-Montanes, J.; Allon, S.J.; Buchheit, K.M.; Vukovic, M.; Derakhshan, T.; Feng, C.; Lai, J.; Hughes, T.K.; Nyquist, S.K.; et al. Human airway mast cells proliferate and acquire distinct inflammation-driven phenotypes during type 2 inflammation. Sci. Immunol. 2021, 6, eabb7221. [Google Scholar] [CrossRef]
- Suurmond, J.; Habets, K.L.L.; Tatum, Z.; Schonkeren, J.J.; Hoen, P.A.; Huizinga, T.W.J.; Laros, J.F.J.; Toes, R.E.M.; Kurreeman, F. Repeated FcepsilonRI triggering reveals modified mast cell function related to chronic allergic responses in tissue. J. Allergy Clin. Immunol. 2016, 138, 869–880. [Google Scholar] [CrossRef]
- Plum, T.; Wang, X.; Rettel, M.; Krijgsveld, J.; Feyerabend, T.B.; Rodewald, H.-R. Human Mast Cell Proteome Reveals Unique Lineage, Putative Functions, and Structural Basis for Cell Ablation. Immunity 2020, 52, 404–416.e5. [Google Scholar] [CrossRef]
- Motakis, E.; Guhl, S.; Ishizu, Y.; Itoh, M.; Kawaji, H.; de Hoon, M.; Lassmann, T.; Carninci, P.; Hayashizaki, Y.; Zuberbier, T.; et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood 2014, 123, e58–e67. [Google Scholar] [CrossRef]
- Guhl, S.; Tapkenhinrichs, S.; Smorodchenko, A.; Grützkau, A.; Henz, B.M.; Zuberbier, T.; Hartmann, K. Ultraviolet Irradiation Induces Apoptosis in Human Immature, But Not in Skin Mast Cells. J. Investig. Dermatol. 2003, 121, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Baranes, D.; Razin, E. Protein kinase C regulates proliferation of mast cells and the expression of the mRNAs of fos and jun proto-oncogenes during activation by IgE-Ag or calcium ionophore A23187. Blood 1991, 78, 2354–2364. [Google Scholar] [CrossRef]
- Tsai, M.; Tam, S.Y.; Galli, S.J. Distinct patterns of early response gene expression and proliferation in mouse mast cells stim-ulated by stem cell factor, interleukin-3, or IgE and antigen. Eur. J. Immunol. 1993, 23, 867–872. [Google Scholar] [CrossRef]
- Razin, E.; Szallasi, Z.; Kazanietz, M.G.; Blumberg, P.M.; Rivera, J. Protein kinases C-beta and C-epsilon link the mast cell high-affinity receptor for IgE to the expression of c-fos and c-jun. Proc. Natl. Acad. Sci. USA 1994, 91, 7722–7726. [Google Scholar] [CrossRef] [PubMed]
- Serve, H.; Yee, N.S.; Stella, G.; Sepp-Lorenzino, L.; Tan, J.C.; Besmer, P. Differential roles of PI3-kinase and Kit tyrosine 821 in Kit receptor-mediated proliferation, survival and cell adhesion in mast cells. EMBO J. 1995, 14, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Tuckerman, J.; Nechushtan, H.; Schutz, G.; Razin, E.; Angel, P. c-Fos as a regulator of degranulation and cytokine production in FcepsilonRI-activated mast cells. J. Immunol. 2004, 173, 2571–2577. [Google Scholar] [CrossRef]
- Lorentz, A.; Wilke, M.; Sellge, G.; Worthmann, H.; Klempnauer, J.; Manns, M.P.; Bischoff, S.C. IL-4-induced priming of human intestinal mast cells for enhanced survival and Th2 cytokine generation is reversible and associated with increased activity of ERK1/2 and c-Fos. J. Immunol. 2005, 174, 6751–6756. [Google Scholar] [CrossRef]
- Li, B.; Power, M.R.; Lin, T.-J. De novo synthesis of early growth response factor-1 is required for the full responsiveness of mast cells to produce TNF and IL-13 by IgE and antigen stimulation. Blood 2006, 107, 2814–2820. [Google Scholar] [CrossRef]
- Li, B.; Berman, J.; Tang, J.-T.; Lin, T.-J. The Early Growth Response Factor-1 Is Involved in Stem Cell Factor (SCF)-induced Interleukin 13 Production by Mast Cells, but Is Dispensable for SCF-dependent Mast Cell Growth. J. Biol. Chem. 2007, 282, 22573–22581. [Google Scholar] [CrossRef]
- Yang, Y.J.; Chen, W.; Edgar, A.; Li, B.; Molkentin, J.D.; Berman, J.N.; Lin, T.-J. Rcan1 negatively regulates Fc epsilonRI-mediated signaling and mast cell function. J. Exp. Med. 2009, 206, 195–207. [Google Scholar] [CrossRef]
- Zhang, L.; Paine, C.; Dip, R. Selective regulation of nuclear orphan receptors 4A by adenosine receptor subtypes in human mast cells. J. Cell Commun. Signal. 2010, 4, 173–183. [Google Scholar] [CrossRef]
- Lundequist, A.; Calounova, G.; Wensman, H.; Rönnberg, E.; Pejler, G. Differential regulation of Nr4a subfamily nuclear receptors following mast cell activation. Mol. Immunol. 2011, 48, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, Y.; MacNeil, A.J.; Junkins, R.D.; Berman, J.N.; Lin, T.-J. Calcineurin–Rcan1 Interaction Contributes to Stem Cell Factor–Mediated Mast Cell Activation. J. Immunol. 2013, 191, 5885–5894. [Google Scholar] [CrossRef]
- Wang, H.N.; Ji, K.; Zhang, L.-N.; Xie, C.-C.; Li, W.-Y.; Zhao, Z.-F.; Chen, J.J. Inhibition of c-Fos expression attenuates IgE-mediated mast cell activation and allergic inflammation by coun-teracting an inhibitory AP1/Egr1/IL-4 axis. J. Transl. Med. 2021, 19, 261. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.S.; Hamey, F.; Pijuan-Sala, B.; Shepherd, M.; Lau, W.; Shaw, S.; Weinreb, C.; Wolock, S.; Hannah, R.; Diamanti, E.; et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood J. Am. Soc. Hematol. 2018, 131, e1–e11. [Google Scholar] [CrossRef] [PubMed]
- Tusi, B.K.; Wolock, S.L.; Weinreb, C.; Hwang, Y.; Hidalgo, D.; Zilionis, R.; Waisman, A.; Huh, J.R.; Klein, A.M.; Socolovsky, M. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 2018, 555, 54–60. [Google Scholar] [CrossRef]
- Inclan-Rico, J.M.; Hernandez, C.M.; Henry, E.K.; Federman, H.G.; Sy, C.B.; Ponessa, J.J.; Lemenze, A.D.; Joseph, N.; Soteropoulos, P.; Beaulieu, A.M.; et al. Trichinella spiralis-induced mastocytosis and erythropoiesis are simultaneously supported by a bipotent mast cell/erythrocyte precursor cell. PLOS Pathog. 2020, 16, e1008579. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.-M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Dahlin, J.S.; Maurer, M.; Metcalfe, D.D.; Pejler, G.; Sagi-Eisenberg, R.; Nilsson, G. The ingenious mast cell: Contemporary insights into mast cell behavior and function. Allergy 2021, 77, 83–99. [Google Scholar] [CrossRef]
- Babina, M.; Kirchhof, L.; Guhl, S.; Franke, R.; Zuberbier, T.; Henz, B.M.; Gombart, A.F. The transcription factor profile of human mast cells in comparison with monocytes and granulocytes. Cell. Mol. Life Sci. 2005, 62, 214–226. [Google Scholar] [CrossRef]
- Calero-Nieto, F.J.; Ng, F.S.; Wilson, N.K.; Hannah, R.; Moignard, V.; Leal-Cervantes, A.I.; Jimenez-Madrid, I.; Diamanti, E.; Wernisch, L.; Göttgens, B. Key regulators control distinct transcriptional programmes in blood progenitor and mast cells. EMBO J. 2014, 33, 1212–1226. [Google Scholar] [CrossRef]
- Isogai, R.; Takahashi, M.; Aisu, K.; Horiuti, Y.; Aragane, Y.; Kawada, A.; Tezuka, T. The receptor for erythropoietin is present on cutaneous mast cells. Arch. Dermatol. Res. 2006, 297, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, T.; Ehrhardt, S.; Cerny, D.; Hildebrand, D.; Klein, S.; Heeg, K.; Kubatzky, K.F. Erythropoietin acts as an anti-inflammatory signal on murine mast cells. Mol. Immunol. 2015, 65, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Boer, A.-K.; Drayer, A.; Vellenga, E. Stem cell factor enhances erythropoietin-mediated transactivation of signal transducer and activator of transcription 5 (STAT5) via the PKA/CREB pathway. Exp. Hematol. 2003, 31, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Luger, S.M.; DeRiel, K.; Abrahm, J.; Calabretta, B.; Gewirtz, A.M. Role of the KIT protooncogene in normal and malignant human hematopoiesis. Proc. Natl. Acad. Sci. USA 1992, 89, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Peschle, C.; Testa, U.; Valtieri, M.; Gabbianelli, M.; Pelosi, E.; Montesoro, E.; Sposi, N.M.; Fossati, C.; Camagna, A.; Caré, A. Stringently purified human hematopoietic progenitors/stem cells: Analysis of cellular/molecular mechanisms underlying early hematopoiesis. STEM CELLS 1993, 11, 356–370. [Google Scholar] [CrossRef]
- Sui, X.; Tsuji, K.; Tajima, S.; Tanaka, R.; Muraoka, K.; Ebihara, Y.; Ikebuchi, K.; Yasukawa, K.; Taga, T.; Kishimoto, T.; et al. Erythropoietin-independent erythrocyte production: Signals through gp130 and c-kit dramatically promote eryth-ropoiesis from human CD34+ cells. J. Exp. Med. 1996, 183, 837–845. [Google Scholar] [CrossRef]
- Kapur, R.; Majumdar, M.; Xiao, X.; McAndrews-Hill, M.; Schindler, K.; A Williams, D. Signaling through the interaction of membrane-restricted stem cell factor and c-kit receptor tyrosine kinase: Genetic evidence for a differential role in erythropoiesis. Blood 1998, 91, 879–889. [Google Scholar] [CrossRef]
- Pharr, P.N.; Hofbauer, A.; E Worthington, R.; Longmore, G.D. Residual erythroid progenitors in W/W mice respond to erythropoietin in the absence of steel factor signals. Int. J. Hematol. 2000, 72, 178–185. [Google Scholar] [CrossRef]
- Munugalavadla, V.; Kapur, R. Role of c-Kit and erythropoietin receptor in erythropoiesis. Crit. Rev. Oncol. Hematol. 2005, 54, 63–75. [Google Scholar] [CrossRef]
- Gregory, R.C.; Jiang, N.; Todokoro, K.; Crouse, J.; E Pacifici, R.; Wojchowski, D.M. Erythropoietin receptor and STAT5-specific pathways promote SKT6 cell hemoglobinization. Blood 1998, 92, 1104–1118. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, G.; Shvartsman, S.Y.; Paroush, Z. The Capicua repressor—A general sensor of RTK signaling in development and disease. J. Cell Sci. 2012, 125, 1383–1391. [Google Scholar] [CrossRef]
- Simón-Carrasco, L.; Graña, O.; Salmón, M.; Jacob, H.K.; Gutierrez, A.; Jiménez, G.; Drosten, M.; Barbacid, M. Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev. 2017, 31, 1456–1468. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, V.G.; Firme, M.; Song, J.; Chan, S.Y.; Lee, M.H.; Yip, S.; Chittaranjan, S.; A Marra, M. Comparative transcriptome analysis of isogenic cell line models and primary cancers links capicua (CIC) loss to activation of the MAPK signalling cascade. J. Pathol. 2017, 242, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y. Regulation and function of capicua in mammals. Exp. Mol. Med. 2020, 52, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Pedersen, M.; Bengtsson, S.; Rönnstrand, L. Grb2 mediates negative regulation of stem cell factor receptor/c-Kit signaling by recruitment of Cbl. Exp. Cell Res. 2007, 313, 3935–3942. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016, 62, 37–49. [Google Scholar] [CrossRef]
- Fowler, T.; Sen, R.; Roy, A.L. Regulation of Primary Response Genes. Mol. Cell 2011, 44, 348–360. [Google Scholar] [CrossRef]

| Gene | Forward 5′-3′ | Reverse 5′-3′ |
|---|---|---|
| FOS | AGTGACCGTGGGAATGAAGT | GCTTCAACGCAGACTACGAG |
| NR4A2 | TTCTGTAACCCTCCTAGCCC | AGCATGGCCAAACATTTCCC |
| JUNB | GCCCGGATGTGCACTAAAAT | GACCAGAAAAGTAGCTGCCG |
| EGR1 | GCCCGGATGTGCACTAAAAT | GACCAGAAAAGTAGCTGCCG |
| RHEX | TACTGAGAGACGAGGTGCCA | AGTTGATGGCGGTGAGGAAG |
| HPRT | GCCTCCCATCTCCTTCATCA | CCTGGCGTCGTGATTAGTGA |
| PPIB * | AAGATGTCCCTGTGCCCTAC | ATGGCAAGCATGTGGTGTTT |
| GAPDH | ATCTCGCTCCTGGAAGATGG | AGGTCGGAGTCAACGGATTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franke, K.; Bal, G.; Li, Z.; Zuberbier, T.; Babina, M. Clorfl86/RHEX Is a Negative Regulator of SCF/KIT Signaling in Human Skin Mast Cells. Cells 2023, 12, 1306. https://doi.org/10.3390/cells12091306
Franke K, Bal G, Li Z, Zuberbier T, Babina M. Clorfl86/RHEX Is a Negative Regulator of SCF/KIT Signaling in Human Skin Mast Cells. Cells. 2023; 12(9):1306. https://doi.org/10.3390/cells12091306
Chicago/Turabian StyleFranke, Kristin, Gürkan Bal, Zhuoran Li, Torsten Zuberbier, and Magda Babina. 2023. "Clorfl86/RHEX Is a Negative Regulator of SCF/KIT Signaling in Human Skin Mast Cells" Cells 12, no. 9: 1306. https://doi.org/10.3390/cells12091306






