Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Isogenic Cell Lines
2.2. Cell Culture and 5-Aza-2-Deoxycytidine Treatment
2.3. Quantitative Reverse Transcriptase Polymerase Chain Reaction
2.4. Pyrosequencing
2.5. Bioinformatics Analysis
2.6. Electrophoretic Mobility Super-Shift Assay
2.7. Chromatin Immunoprecipitation and qPCR
2.8. Transfection of Small Interference RNA
2.9. Western Blotting Analysis
2.10. Cell Migration Assay
2.11. Cell Proliferation Assay
2.12. Cell Apoptosis Assay
2.13. Statistical Analyses
3. Results
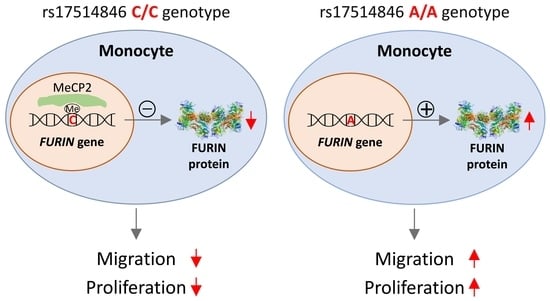
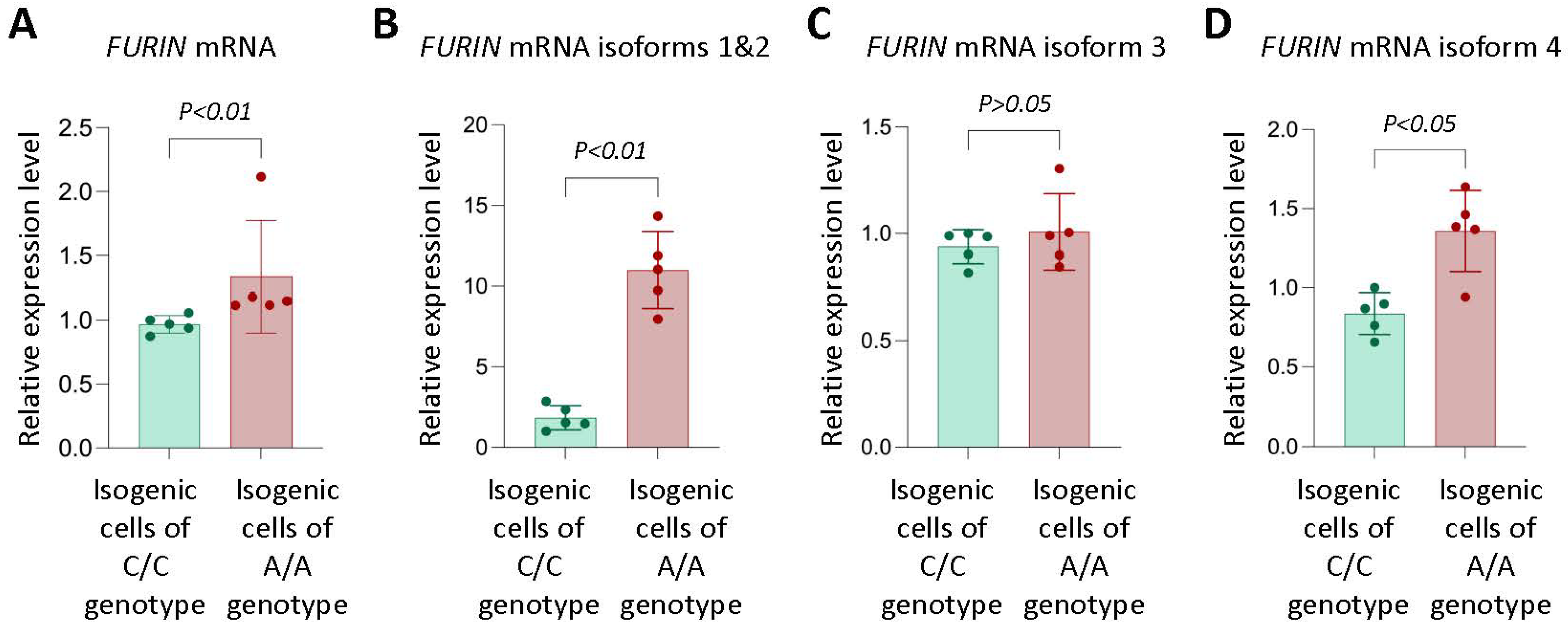
3.1. Allelic Effect of rs17514846 on FURIN Expression
3.2. Allelic Effect of rs17514846 on DNA Methylation
3.3. DNA Methylation Inhibition Increased FURIN Expression
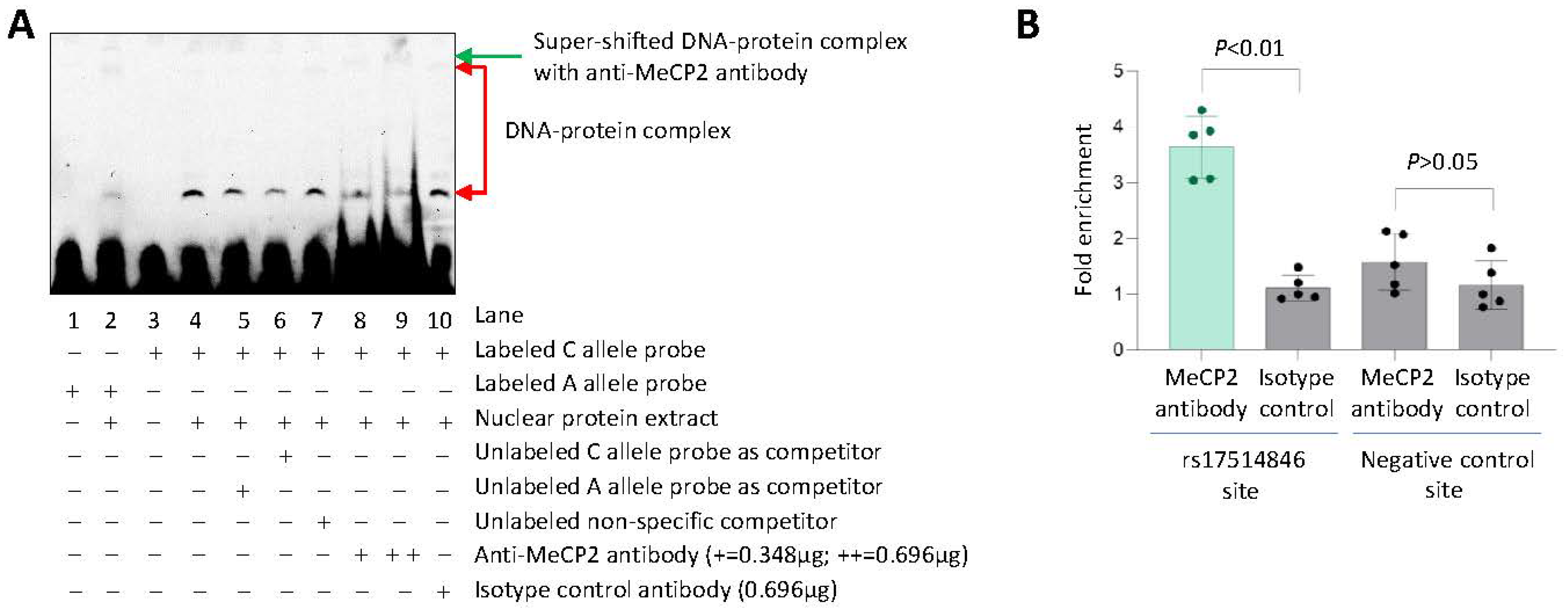
3.4. Allele-Differential Binding of the Transcription Factor MeCP2
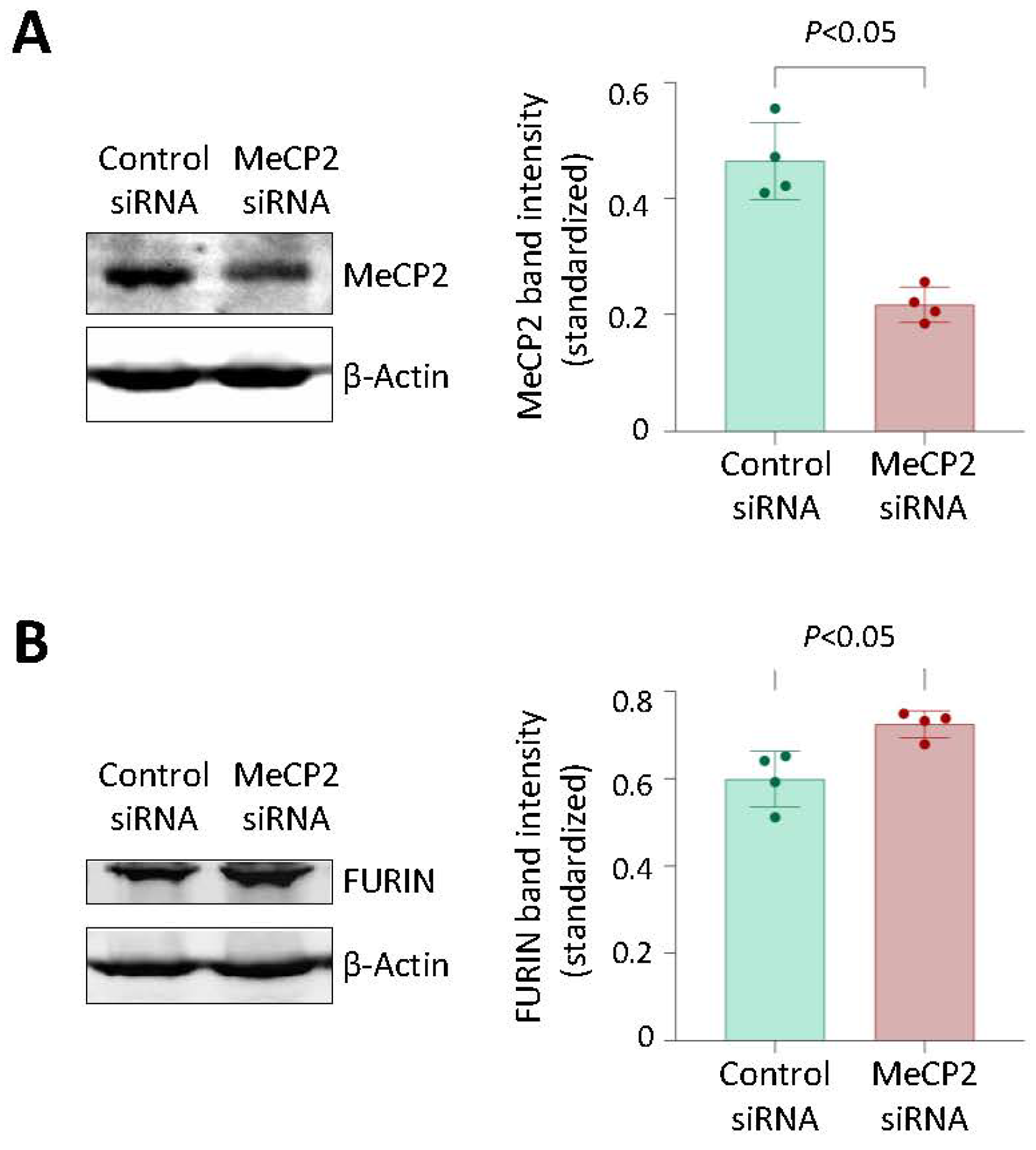
3.5. Knockdown of MeCP2 Increased FURIN Expression
3.6. MeCP2 Knockdown Increased Monocyte Migration and Proliferation, and These Effects were Attenuated by a FURIN Inhibitor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CARDIoGRAMplusC4D_Consortium; Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [PubMed]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Howson, J.M.M.; Zhao, W.; Barnes, D.R.; Ho, W.K.; Young, R.; Paul, D.S.; Waite, L.L.; Freitag, D.F.; Fauman, E.B.; Salfati, E.L.; et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017, 49, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yang, W.; Wu, J.; Chen, B.; Yang, X.; Chen, J.; McVey, D.G.; Andreadi, C.; Gong, P.; Webb, T.R.; et al. Influence of a Coronary Artery Disease-Associated Genetic Variant on FURIN Expression and Effect of Furin on Macrophage Behavior. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1837–1844. [Google Scholar] [CrossRef]
- Benjannet, S.; Rhainds, D.; Hamelin, J.; Nassoury, N.; Seidah, N.G. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: Functional consequences of natural mutations and post-translational modifications. J. Biol. Chem. 2006, 281, 30561–30572. [Google Scholar] [CrossRef]
- Yana, I.; Weiss, S.J. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell 2000, 11, 2387–2401. [Google Scholar] [CrossRef]
- Dubois, C.M.; Blanchette, F.; Laprise, M.H.; Leduc, R.; Grondin, F.; Seidah, N.G. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am. J. Pathol. 2001, 158, 305–316. [Google Scholar] [CrossRef]
- Duguay, S.J.; Milewski, W.M.; Young, B.D.; Nakayama, K.; Steiner, D.F. Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J. Biol. Chem. 1997, 272, 6663–6670. [Google Scholar] [CrossRef]
- Bresnahan, P.A.; Leduc, R.; Thomas, L.; Thorner, J.; Gibson, H.L.; Brake, A.J.; Barr, P.J.; Thomas, G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J. Cell Biol. 1990, 111, 2851–2859. [Google Scholar] [CrossRef]
- Lissitzky, J.C.; Luis, J.; Munzer, J.S.; Benjannet, S.; Parat, F.; Chretien, M.; Marvaldi, J.; Seidah, N.G. Endoproteolytic processing of integrin pro-alpha subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem. J. 2000, 346 Pt 1, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Anders, A.; Gilbert, S.; Garten, W.; Postina, R.; Fahrenholz, F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001, 15, 1837–1839. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Broder, C.; Helmstetter, A.; Schmidt, S.; Yan, I.; Muller, M.; Schmidt-Arras, D.; Becker-Pauly, C.; Koch-Nolte, F.; Mittrucker, H.W.; et al. Short-term TNFalpha shedding is independent of cytoplasmic phosphorylation or furin cleavage of ADAM17. Biochim. Biophys. Acta 2013, 1833, 3355–3367. [Google Scholar] [CrossRef]
- Wang, P.; Tortorella, M.; England, K.; Malfait, A.M.; Thomas, G.; Arner, E.C.; Pei, D. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J. Biol. Chem. 2004, 279, 15434–15440. [Google Scholar] [CrossRef]
- Somerville, R.P.; Longpre, J.M.; Apel, E.D.; Lewis, R.M.; Wang, L.W.; Sanes, J.R.; Leduc, R.; Apte, S.S. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J. Biol. Chem. 2004, 279, 35159–35175. [Google Scholar] [CrossRef]
- Turpeinen, H.; Raitoharju, E.; Oksanen, A.; Oksala, N.; Levula, M.; Lyytikainen, L.P.; Jarvinen, O.; Creemers, J.W.; Kahonen, M.; Laaksonen, R.; et al. Proprotein convertases in human atherosclerotic plaques: The overexpression of FURIN and its substrate cytokines BAFF and APRIL. Atherosclerosis 2011, 219, 799–806. [Google Scholar] [CrossRef]
- Sluijter, J.P.; Verloop, R.E.; Pulskens, W.P.; Velema, E.; Grimbergen, J.M.; Quax, P.H.; Goumans, M.J.; Pasterkamp, G.; de Kleijn, D.P. Involvement of furin-like proprotein convertases in the arterial response to injury. Cardiovasc. Res. 2005, 68, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Stawowy, P.; Marcinkiewicz, J.; Graf, K.; Seidah, N.; Chretien, M.; Fleck, E.; Marcinkiewicz, M. Selective expression of the proprotein convertases furin, pc5, and pc7 in proliferating vascular smooth muscle cells of the rat aorta in vitro. J. Histochem. Cytochem. 2001, 49, 323–332. [Google Scholar] [CrossRef]
- Stawowy, P.; Kallisch, H.; Borges Pereira Stawowy, N.; Stibenz, D.; Veinot, J.P.; Grafe, M.; Seidah, N.G.; Chretien, M.; Fleck, E.; Graf, K. Immunohistochemical localization of subtilisin/kexin-like proprotein convertases in human atherosclerosis. Virchows Arch. 2005, 446, 351–359. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in atherosclerosis. Nature 2002, 420, 868–874. [Google Scholar] [CrossRef]
- Yakala, G.K.; Cabrera-Fuentes, H.A.; Crespo-Avilan, G.E.; Rattanasopa, C.; Burlacu, A.; George, B.L.; Anand, K.; Mayan, D.C.; Corliano, M.; Hernandez-Resendiz, S.; et al. FURIN Inhibition Reduces Vascular Remodeling and Atherosclerotic Lesion Progression in Mice. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Tang, J.N.; Han, L.; Liu, X.D.; Shen, Y.L.; Zhang, C.Y.; Liu, X.B. Elevated FURIN levels in predicting mortality and cardiovascular events in patients with acute myocardial infarction. Metabolism 2020, 111, 154323. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Cross, S.; Bird, A. Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp. 1998, 214, 6–16; Discussion 16–21, 46–50. [Google Scholar]
- Fuks, F.; Hurd, P.J.; Wolf, D.; Nan, X.; Bird, A.P.; Kouzarides, T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003, 278, 4035–4040. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Perez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Zheng, R.; Wan, C.; Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Chen, C.H.; Brown, M.; Zhang, X.; Meyer, C.A.; et al. Cistrome Data Browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019, 47, D729–D735. [Google Scholar] [CrossRef]
- Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Zheng, R.; Zang, C.; Zhu, M.; Wu, J.; Shi, X.; Taing, L.; et al. Cistrome Data Browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017, 45, D658–D662. [Google Scholar] [CrossRef]
- Ayoubi, T.A.; Creemers, J.W.; Roebroek, A.J.; Van de Ven, W.J. Expression of the dibasic proprotein processing enzyme furin is directed by multiple promoters. J. Biol. Chem. 1994, 269, 9298–9303. [Google Scholar] [CrossRef] [PubMed]
- Tcheandjieu, C.; Zhu, X.; Hilliard, A.T.; Clarke, S.L.; Napolioni, V.; Ma, S.; Lee, K.M.; Fang, H.; Chen, F.; Lu, Y.; et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat. Med. 2022, 28, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Atri, D.S.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W.; et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 2022, 54, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Angelakopoulou, A.; Shah, T.; Sofat, R.; Shah, S.; Berry, D.J.; Cooper, J.; Palmen, J.; Tzoulaki, I.; Wong, A.; Jefferis, B.J.; et al. Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. Eur. Heart J. 2012, 33, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; Konig, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated with Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef]
- McMahon, S.; Grondin, F.; McDonald, P.P.; Richard, D.E.; Dubois, C.M. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: Impact on the bioactivation of proproteins. J. Biol. Chem. 2005, 280, 6561–6569. [Google Scholar] [CrossRef]
- Laprise, M.H.; Grondin, F.; Cayer, P.; McDonald, P.P.; Dubois, C.M. Furin gene (fur) regulation in differentiating human megakaryoblastic Dami cells: Involvement of the proximal GATA recognition motif in the P1 promoter and impact on the maturation of furin substrates. Blood 2002, 100, 3578–3587. [Google Scholar] [CrossRef]
- Blanchette, F.; Rudd, P.; Grondin, F.; Attisano, L.; Dubois, C.M. Involvement of Smads in TGFbeta1-induced furin (fur) transcription. J. Cell Physiol. 2001, 188, 264–273. [Google Scholar] [CrossRef]
- Pesu, M.; Muul, L.; Kanno, Y.; O’Shea, J.J. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood 2006, 108, 983–985. [Google Scholar] [CrossRef]
- He, Z.; Khatib, A.M.; Creemers, J.W.M. The proprotein convertase furin in cancer: More than an oncogene. Oncogene 2022, 41, 1252–1262. [Google Scholar] [CrossRef]
- Couture, F.; Kwiatkowska, A.; Dory, Y.L.; Day, R. Therapeutic uses of furin and its inhibitors: A patent review. Expert Opin. Ther. Pat. 2015, 25, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhang, X.; Su, J.; Jeong, M.; Gundry, M.C.; Huang, Y.H.; Zhou, Y.; Li, W.; Goodell, M.A. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017, 8, 16026. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Cao, J.; McVey, D.G.; Ye, S. Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus. Cells 2023, 12, 1681. https://doi.org/10.3390/cells12131681
Yang W, Cao J, McVey DG, Ye S. Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus. Cells. 2023; 12(13):1681. https://doi.org/10.3390/cells12131681
Chicago/Turabian StyleYang, Wei, Junjun Cao, David G. McVey, and Shu Ye. 2023. "Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus" Cells 12, no. 13: 1681. https://doi.org/10.3390/cells12131681
APA StyleYang, W., Cao, J., McVey, D. G., & Ye, S. (2023). Allele-Specific Epigenetic Regulation of FURIN Expression at a Coronary Artery Disease Susceptibility Locus. Cells, 12(13), 1681. https://doi.org/10.3390/cells12131681