Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Subcellular Fractionation of Rat Liver
2.3. Preparation of Endolysosomal Membrane-Enriched Fractions
2.4. Co-Immunoprecipitation
2.5. Western Blotting
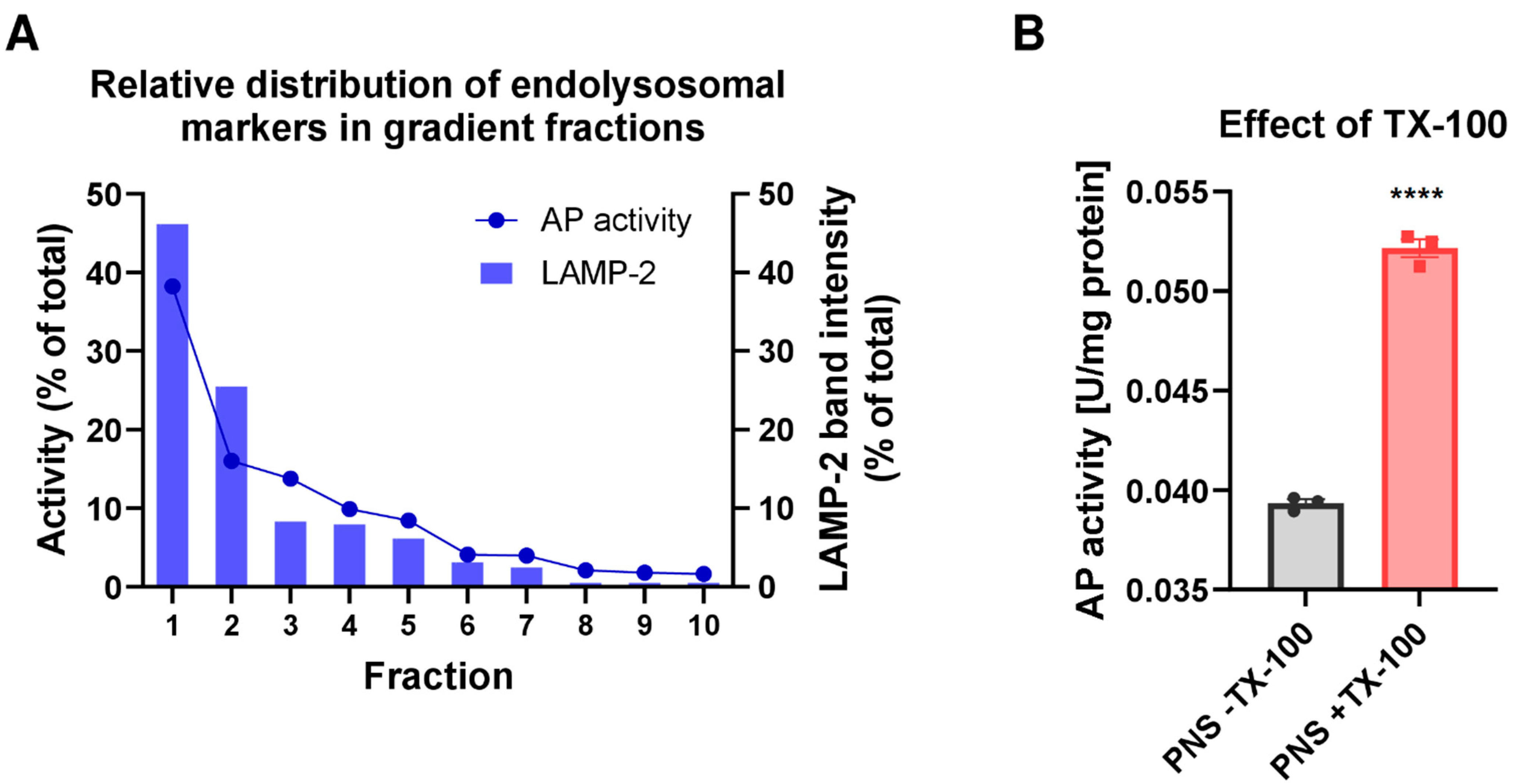
2.6. Assessment of Gradient Fractionation Efficiency and Distribution of Pgp in the Marker Organelle Enriched Fractions
2.7. Acid Phosphatase (AP) Assay
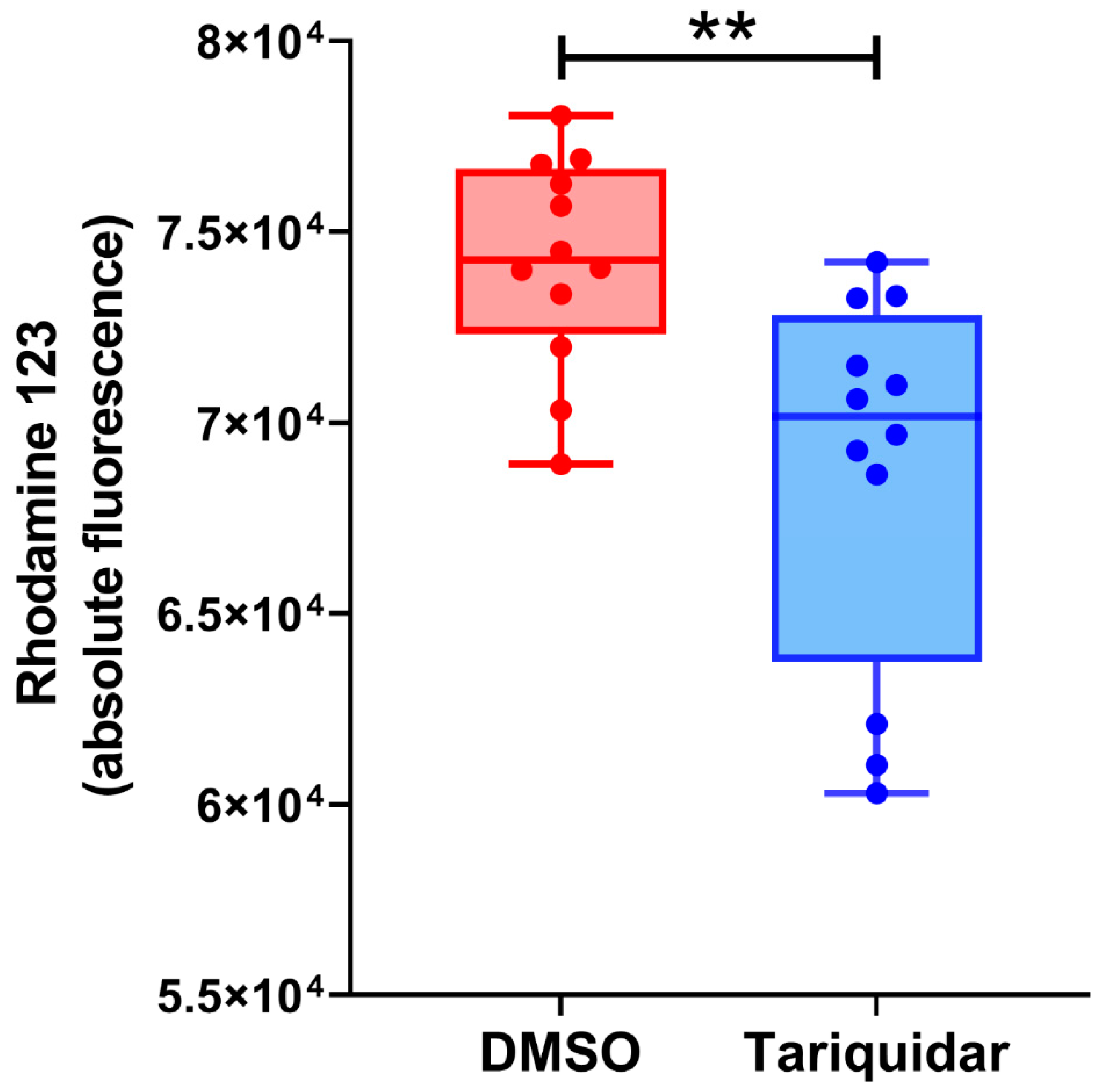
2.8. Rhodamine 123 Uptake Assay Using the Crude Lysosomal Fraction (CLF) of Rat Liver
2.9. Electron Microscopy of the Subcellular Fractions and Immunopurified Vesicles
2.10. Ultrastructure of the Rat Liver
2.11. Electron Microscopy of hCMEC/D3 Cells with vs. without Exposure to Doxorubicin
2.12. Statistics
3. Results
3.1. Biochemical Characterization of the Endolysosome-Enriched Fraction Obtained by Using the Basic Fractionation Protocol
3.2. Biochemical Analysis of Pgp and Organelle Marker Distribution in Subcellular Fractions following an Improved Fractionation Protocol
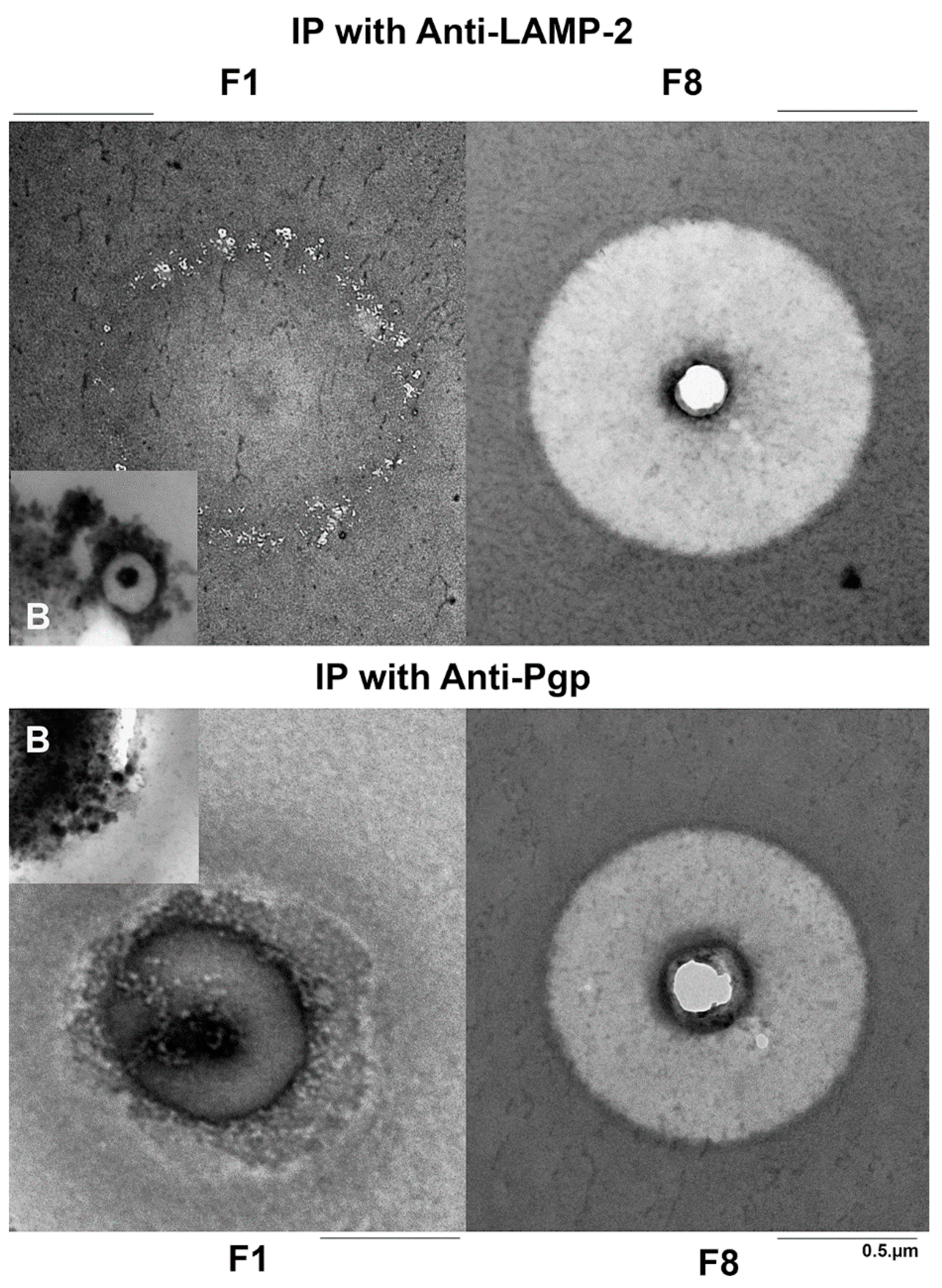
3.3. Ultrastructural Analyses of the Subcellular Fractions by Negative Staining
3.4. Ultrastructural Comparison of Anti-LAMP-2 and Anti-Pgp Immunopurified Vesicles
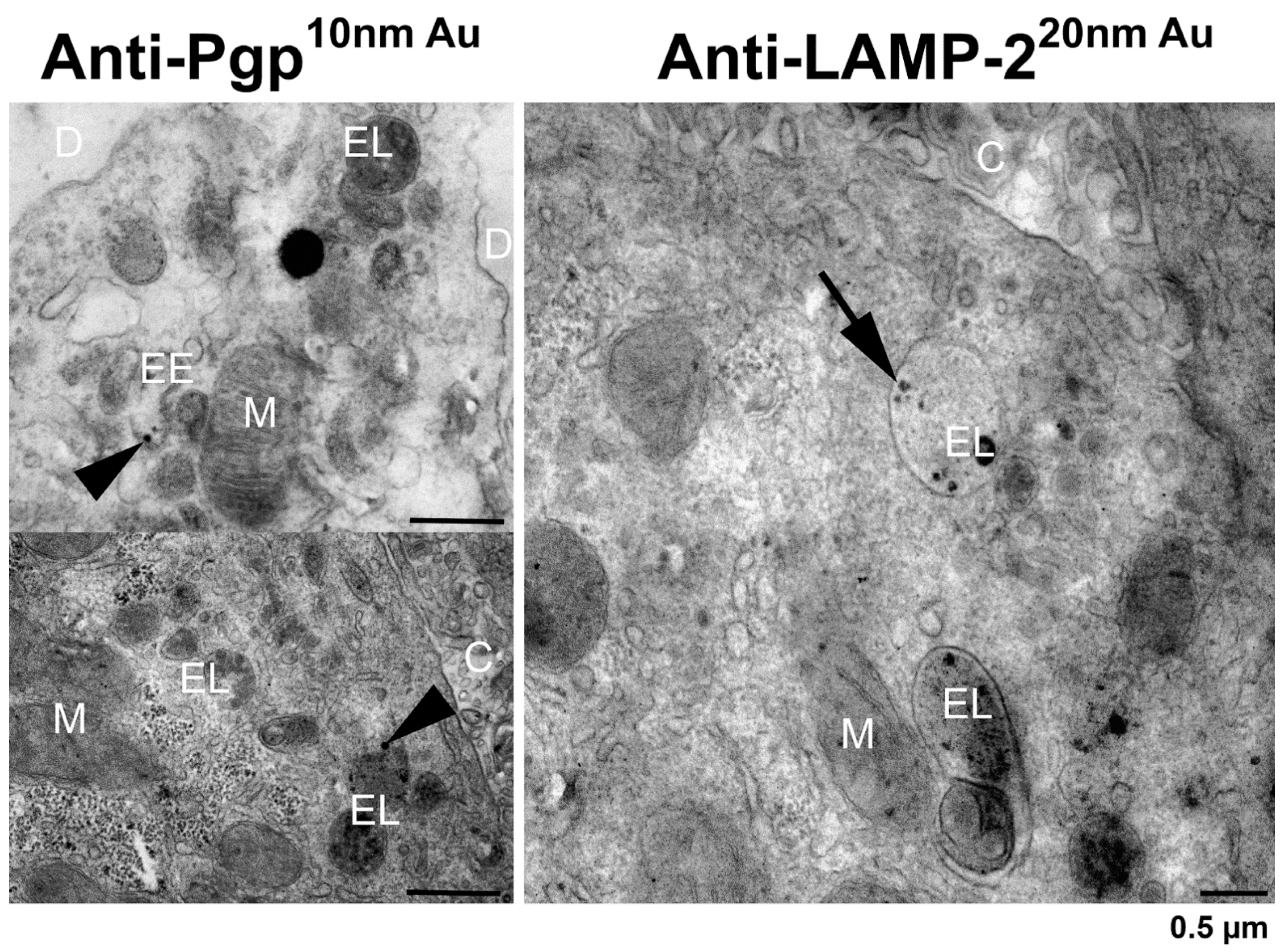
3.5. In Situ Localization of Pgp and LAMP-2 in Rat Liver by Pre-embedding Immunogold Electron Microscopy
3.6. Summary of the Biochemical and Ultrastructural Data
3.7. Electron Microscopy of hCMEC/D3-MDR1-EGFP Cells with vs. without Exposure to Doxorubicin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ling, V. Multidrug resistance: Molecular mechanisms and clinical relevance. Cancer Chemother. Pharmacol. 1997, 40, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Seelig, A. P-Glycoprotein: One Mechanism, Many Tasks and the Consequences for Pharmacotherapy of Cancers. Front. Oncol. 2020, 10, 576559. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, A.H. P-Glycoprotein, a gatekeeper in the blood-brain barrier. Adv. Drug Deliv. Rev. 1999, 36, 179–194. [Google Scholar] [CrossRef]
- Löscher, W.; Potschka, H. Drug resistance in brain diseases and the role of drug efflux transporters. Nat. Rev. Neurosci. 2005, 6, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Schinkel, A.H. P-glycoprotein ABCB1: A major player in drug handling by mammals. J. Clin. Investig. 2013, 123, 4131–4133. [Google Scholar] [CrossRef]
- Löscher, W.; Gericke, B. Novel Intrinsic Mechanisms of Active Drug Extrusion at the Blood-Brain Barrier: Potential Targets for Enhancing Drug Delivery to the Brain? Pharmaceutics 2020, 12, 966. [Google Scholar] [CrossRef]
- Koenig, J.; Müller, F.; Fromm, M.F. Transporters and Drug-Drug Interactions: Important Determinants of Drug Disposition and Effects. Pharmacol. Rev. 2013, 65, 944–966. [Google Scholar] [CrossRef]
- Yamagishi, T.; Sahni, S.; Sharp, D.M.; Arvind, A.; Jansson, P.J.; Richardson, D.R. P-glycoprotein Mediates Drug Resistance via a Novel Mechanism Involving Lysosomal Sequestration. J. Biol. Chem. 2013, 288, 31761–31771. [Google Scholar] [CrossRef]
- Rajagopal, A.; Simon, S.M. Subcellular Localization and Activity of Multidrug Resistance Proteins. Mol. Biol. Cell 2003, 14, 3389–3399. [Google Scholar] [CrossRef]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomes as mediators of drug resistance in cancer. Drug Resist. Updates 2016, 24, 23–33. [Google Scholar] [CrossRef]
- Seebacher, N.; Lane, D.; Richardson, D.; Jansson, P. Turning the gun on cancer: Utilizing lysosomal P-glycoprotein as a new strategy to overcome multi-drug resistance. Free Radic. Biol. Med. 2016, 96, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Stefan, S.M.; Jansson, P.; Kalinowski, D.S.; Anjum, R.; Dharmasivam, M.; Richardson, D.R. The growing evidence for targeting P-glycoprotein in lysosomes to overcome resistance. Future Med. Chem. 2020, 12, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Noack, A.; Gericke, B.; von Köckritz-Blickwede, M.; Menze, A.; Noack, S.; Gerhauser, I.; Osten, F.; Naim, H.Y.; Löscher, W. A novel mechanism of drug extrusion by brain endothelial cells via lysosomal drug trapping and disposal by neutrophils. Proc. Natl. Acad. Sci. USA 2018, 115, E9590–E9599. [Google Scholar] [CrossRef] [PubMed]
- Szakacs, G.; Abele, R. An inventory of lysosomal ABC transporters. FEBS Lett. 2020, 594, 3965–3985. [Google Scholar] [CrossRef]
- Fu, D.; Arias, I.M. Intracellular trafficking of P-glycoprotein. Int. J. Biochem. Cell Biol. 2012, 44, 461–464. [Google Scholar] [CrossRef]
- Fu, D. Where is it and How Does it Get There—Intracellular Localization and Traffic of P-glycoprotein. Front. Oncol. 2013, 3, 321. [Google Scholar] [CrossRef]
- Liu-Kreyche, P.; Shen, H.; Marino, A.M.; Iyer, R.A.; Humphreys, W.G.; Lai, Y. Lysosomal P-gp-MDR1 Confers Drug Resistance of Brentuximab Vedotin and Its Cytotoxic Payload Monomethyl Auristatin E in Tumor Cells. Front. Pharmacol. 2019, 10, 749. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef]
- Elferink, R.P.J.O.; Paulusma, C. Function and pathophysiological importance of ABCB4 (MDR3 P-glycoprotein). Pflug. Arch. 2006, 453, 601–610. [Google Scholar] [CrossRef]
- Graham, J.M. Isolation of Lysosomes from Tissues and Cells by Differential and Density Gradient Centrifugation. Curr. Protoc. Cell Biol. 2000, 7, 3.5.1–3.5.21. [Google Scholar] [CrossRef]
- Aguado, C.; Pérez-Jiménez, E.; Lahuerta, M.; Knecht, E. Isolation of Lysosomes from Mammalian Tissues and Cultured Cells. Methods Mol. Biol. 2016, 1449, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, J.; Tchikov, V.; Hennig, L.; Lucius, R.; Schütze, S. A toolbox for the immunomagnetic purification of signaling organelles. Traffic 2019, 20, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Kaade, E.; Muntel, J.; Bruderer, R.; Reiter, L.; Thelen, M.; Winter, D. Systematic Comparison of Strategies for the Enrichment of Lysosomes by Data Independent Acquisition. J. Proteome Res. 2019, 19, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Klumperman, J.; Raposo, G. The Complex Ultrastructure of the Endolysosomal System. Cold Spring Harb. Perspect. Biol. 2014, 6, a016857. [Google Scholar] [CrossRef] [PubMed]
- Bright, N.A.; Davis, L.J.; Luzio, J.P. Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity. Curr. Biol. 2016, 26, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- De Araujo, M.E.; Liebscher, G.; Hess, M.W.; Huber, L.A. Lysosomal size matters. Traffic 2019, 21, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Montealegre, D.; Pearce, D.A. Defective lysosomal arginine transport in juvenile Batten disease. Hum. Mol. Genet. 2005, 14, 3759–3773. [Google Scholar] [CrossRef]
- Sudhakar, J.N.; Lu, H.-H.; Chiang, H.-Y.; Suen, C.-S.; Hwang, M.-J.; Wu, S.-Y.; Shen, C.-N.; Chang, Y.-M.; Li, F.-A.; Liu, F.-T.; et al. Lumenal Galectin-9-Lamp2 interaction regulates lysosome and autophagy to prevent pathogenesis in the intestine and pancreas. Nat. Commun. 2020, 11, 4286. [Google Scholar] [CrossRef]
- Kartner, N.; Evernden-Porelle, D.; Bradley, G.; Ling, V. Detection of P-glycoprotein in multidrug-resistant cell lines by monoclonal antibodies. Nature 1985, 316, 820–823. [Google Scholar] [CrossRef]
- Okochi, E.; Iwahashi, T.; Tsuruo, T. Monoclonal antibodies specific for P-glycoprotein. Leukemia 1997, 11, 1119–1123. [Google Scholar] [CrossRef][Green Version]
- Elsen, J.M.H.V.D.; Kuntz, D.A.; Hoedemaeker, F.J.; Rose, D.R. Antibody C219 recognizes an α-helical epitope on P-glycoprotein. Proc. Natl. Acad. Sci. USA 1999, 96, 13679–13684. [Google Scholar] [CrossRef] [PubMed]
- Volk, H.; Potschka, H.; Löscher, W. Immunohistochemial localization of P-glycoprotein in rat brain and detection of its increased expression by seizures are sensitive to fixation and staining variables. J. Histochem. Cytochem. 2005, 53, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, K.; Löscher, W. Upregulation of Brain Expression of P-Glycoprotein in MRP2-deficient TR-Rats Resembles Seizure-induced Up-regulation of This Drug Efflux Transporter in Normal Rats. Epilepsia 2007, 48, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Stremmel, W.; Staffer, S.; Gan-Schreier, H.; Wannhoff, A.; Bach, M.; Gauss, A. Phosphatidylcholine passes through lateral tight junctions for paracellular transport to the apical side of the polarized intestinal tumor cell-line CaCo2. Biochim. Et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Noack, A.; Noack, S.; Hoffmann, A.; Maalouf, K.; Buettner, M.; Couraud, P.-O.; Romero, I.A.; Weksler, B.; Alms, D.; Römermann, K.; et al. Drug-Induced Trafficking of P-Glycoprotein in Human Brain Capillary Endothelial Cells as Demonstrated by Exposure to Mitomycin C. PLoS ONE 2014, 9, e88154. [Google Scholar] [CrossRef]
- Noack, A.; Noack, S.; Buettner, M.; Naim, H.Y.; Löscher, W. Intercellular transfer of P-glycoprotein in human blood-brain barrier endothelial cells is increased by histone deacetylase inhibitors. Sci. Rep. 2016, 6, 29253. [Google Scholar] [CrossRef]
- Gericke, B.; Borsdorf, S.; Wienböker, I.; Noack, A.; Noack, S.; Löscher, W. Similarities and differences in the localization, trafficking, and function of P-glycoprotein in MDR1-EGFP-transduced rat versus human brain capillary endothelial cell lines. Fluids Barriers. CNS. 2021, 18, 36. [Google Scholar] [CrossRef]
- Cho, M.; Lee, Z.-W.; Cho, H.-T. ATP-Binding Cassette B4, an Auxin-Efflux Transporter, Stably Associates with the Plasma Membrane and Shows Distinctive Intracellular Trafficking from That of PIN-FORMED Proteins. Plant Physiol. 2012, 159, 642–654. [Google Scholar] [CrossRef]
- Pannese, E.; Luciano, L.; Iurato, S.; Reale, E. Lysosomes in normal and degenerating neuroblasts of the chick embryo spinal ganglia. A cytochemical and quantitative study by electron microscopy. Acta Neuropathol. 1976, 36, 209–220. [Google Scholar] [CrossRef]
- van Meel, E.; Klumperman, J. Imaging and imagination: Understanding the endo-lysosomal system. Histochem. Cell Biol. 2008, 129, 253–266. [Google Scholar] [CrossRef]
- Runquist, E.; Havel, R. Acid hydrolases in early and late endosome fractions from rat liver. J. Biol. Chem. 1991, 266, 22557–22563. [Google Scholar] [CrossRef]
- Braun, M.; Waheed, A.; Von Figura, K. Lysosomal acid phosphatase is transported to lysosomes via the cell surface. EMBO J. 1989, 8, 3633–3640. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yano, S.; Okada, K.; Ishikawa, T.; Himeno, M.; Kato, K. Lysosomal acid phosphatase is transported via endosomes to lysosomes. Biochem. Biophys. Res. Commun. 1990, 166, 1176–1182. [Google Scholar] [CrossRef]
- Pasternak, S.H.; Bagshaw, R.D.; Guiral, M.; Zhang, S.; Ackerley, C.A.; Pak, B.J.; Callahan, J.W.; Mahuran, D.J. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J. Biol. Chem. 2003, 278, 26687–26694. [Google Scholar] [CrossRef]
- Fardel, O.; Le Vee, M.; Jouan, E.; Denizot, C.; Parmentier, Y. Nature and uses of fluorescent dyes for drug transporter studies. Expert Opin. Drug Metab. Toxicol. 2015, 11, 1233–1251. [Google Scholar] [CrossRef]
- Saftig, P.; Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol 2015, 77, 57–80. [Google Scholar] [CrossRef]
- Akasaki, K.; Michihara, A.; Fujiwara, Y.; Mibuka, K.; Tsuji, H. Biosynthetic Transport of a Major Lysosome-Associated Membrane Glycoprotein 2, Lamp-2: A Significant Fraction of Newly Synthesized Lamp-2 Is Delivered to Lysosomes by Way of Early Endosomes. J. Biochem. 1996, 120, 1088–1094. [Google Scholar] [CrossRef]
- Karlsson, K.; Carlsson, S.R. Sorting of lysosomal membrane glycoproteins lamp-1 and lamp-2 into vesicles distinct from mannose 6-phosphate receptor/gamma-adaptin vesicles at the trans-Golgi network. J. Biol. Chem. 1998, 273, 18966–18973. [Google Scholar] [CrossRef]
- Braulke, T.; Bonifacino, J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta 2009, 1793, 605–614. [Google Scholar] [CrossRef]
- Jansson, P.J.; Yamagishi, T.; Arvind, A.; Seebacher, N.; Gutierrez, E.; Stacy, A.; Maleki, S.; Sharp, D.; Sahni, S.; Richardson, D.R. Di-2-pyridylketone 4,4-Dimethyl-3-thiosemicarbazone (Dp44mT) Overcomes Multidrug Resistance by a Novel Mechanism Involving the Hijacking of Lysosomal P-Glycoprotein (Pgp). J. Biol. Chem. 2015, 290, 9588–9603. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, N.; Lane, D.; Jansson, P.J.; Richardson, D.R. Glucose Modulation Induces Lysosome Formation and Increases Lysosomotropic Drug Sequestration via the P-Glycoprotein Drug Transporter. J. Biol. Chem. 2016, 291, 3796–3820. [Google Scholar] [CrossRef] [PubMed]
- Al-Akra, L.; Bae, D.-H.; Sahni, S.; Huang, M.L.; Park, K.C.; Lane, D.; Jansson, P.J.; Richardson, D.R. Tumor stressors induce two mechanisms of intracellular P-glycoprotein–mediated resistance that are overcome by lysosomal-targeted thiosemicarbazones. J. Biol. Chem. 2018, 293, 3562–3587. [Google Scholar] [CrossRef]
- Orlowski, S.; Martin, S.; Escargueil, A. P-glycoprotein and ‘lipid rafts’: Some ambiguous mutual relationships (floating on them, building them or meeting them by chance?). Cell Mol. Life Sci. 2006, 63, 1038–1059. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.-Y.; Tsuda, T.; Horikami, M.; Teraoka, R.; Kitagawa, S.; Terada, T. Bile salt-stimulated phospholipid efflux mediated by ABCB4 localized in nonraft membranes. J. Lipid Res. 2013, 54, 1221–1230. [Google Scholar] [CrossRef]
- Katayama, K.; Kapoor, K.; Ohnuma, S.; Patel, A.; Swaim, W.; Ambudkar, I.S.; Ambudkar, S.V. Revealing the fate of cell surface human P-glycoprotein (ABCB1): The lysosomal degradation pathway. Biochim. Biophys. Acta 2015, 1853, 2361–2370. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.B.; Fox, K.; Lee, P.; Yang, Y.D.; Ling, V. Functional intracellular P-glycoprotein. Int. J. Cancer 1998, 76, 857–864. [Google Scholar] [CrossRef]
- Kipp, H.; Pichetshote, N.; Arias, I.M. Transporters on demand: Intrahepatic pools of canalicular ATP binding cassette transporters in rat liver. J Biol. Chem. 2001, 276, 7218–7224. [Google Scholar] [CrossRef]
- Fu, D.; van Dam, E.M.; Brymora, A.; Duggin, I.G.; Robinson, P.J.; Roufogalis, B.D. The small GTPases Rab5 and RalA regulate intracellular traffic of P-glycoprotein. Biochim. Biophys. Acta 2007, 1773, 1062–1072. [Google Scholar] [CrossRef]
- Cullen, P.J.; Steinberg, F. To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef]
- Chapel, A.; Kieffer-Jaquinod, S.; Sagne, C.; Verdon, Q.; Ivaldi, C.; Mellal, M.; Thirion, J.; Jadot, M.; Bruley, C.; Garin, J.; et al. An Extended Proteome Map of the Lysosomal Membrane Reveals Novel Potential Transporters. Mol. Cell. Proteom. 2013, 12, 1572–1588. [Google Scholar] [CrossRef] [PubMed]
- Della Valle, M.C.; Sleat, D.E.; Zheng, H.; Moore, D.F.; Jadot, M.; Lobel, P. Classification of Subcellular Location by Comparative Proteomic Analysis of Native and Density-shifted Lysosomes. Mol. Cell. Proteom. 2011, 10, M110. [Google Scholar] [CrossRef] [PubMed]
- Thelen, M.; Winter, D.; Braulke, T.; Gieselmann, V. SILAC-Based Comparative Proteomic Analysis of Lysosomes from Mammalian Cells Using LC-MS/MS. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1594, pp. 1–18. [Google Scholar] [CrossRef]
- Lebedeva, I.V.; Pande, P.; Patton, W.F. Sensitive and Specific Fluorescent Probes for Functional Analysis of the Three Major Types of Mammalian ABC Transporters. PLoS ONE 2011, 6, e22429. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Tam, A.; Santi, S.A.; Parissenti, A.M. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in MCF-7 cells. BMC Cancer 2016, 16, 762. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lu, L.; Yan, S.; Yi, H.; Yao, H.; Wu, D.; He, G.; Tao, X.; Deng, X. Autophagy and doxorubicin resistance in cancer. Anti-Cancer Drugs 2018, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Usman, R.M.; Razzaq, F.; Akbar, A.; Farooqui, A.A.; Iftikhar, A.; Latif, A.; Hassan, H.; Zhao, J.; Carew, J.S.; Nawrocki, S.T.; et al. Role and mechanism of autophagy-regulating factors in tumorigenesis and drug resistance. Asia-Pac. J. Clin. Oncol. 2020, 17, 193–208. [Google Scholar] [CrossRef]
- Fader, C.M.; Colombo, M.I. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2008, 16, 70–78. [Google Scholar] [CrossRef]
- Zhang, M.; Schekman, R. Cell biology. Unconventional secretion, unconventional solutions. Science 2013, 340, 559–561. [Google Scholar] [CrossRef]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front. Pharmacol. 2021, 12, 648407. [Google Scholar] [CrossRef]
- Aberuyi, N.; Rahgozar, S.; Dehaghi, Z.K.; Moafi, A.; Masotti, A.; Paolini, A. The translational expression of ABCA2 and ABCA3 is a strong prognostic biomarker for multidrug resistance in pediatric acute lymphoblastic leukemia. OncoTargets Ther. 2017, 10, 3373–3380. [Google Scholar] [CrossRef]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef] [PubMed]
- Sewell, R.B.; Dillon, C.; Grinpukel, S.; Yeomans, N.D.; Smallwood, R.A. Pericanalicular location of hepatocyte lysosomes and effects of fasting: A morphometric analysis. Hepatology 1986, 6, 305–311. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. Lysosomes revisited. Eur. J. Biochem. 1983, 137, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.; LaRusso, N.F. The liver and intracellular digestion: How liver cells eat! Hepatology 1989, 10, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Ballabio, A. Lysosomal Adaptation: How the Lysosome Responds to External Cues. Cold Spring Harb. Perspect. Biol. 2014, 6, a016907. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef]
- Jeger, J.L. Endosomes, lysosomes, and the role of endosomal and lysosomal biogenesis in cancer development. Mol. Biol. Rep. 2020, 47, 9801–9810. [Google Scholar] [CrossRef]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef]
- Heffeter, P.; Pape, V.F.S.; Enyedy, É.A.; Keppler, B.K.; Szakacs, G.; Kowol, C.R. Anticancer Thiosemicarbazones: Chemical Properties, Interaction with Iron Metabolism, and Resistance Development. Antioxid. Redox. Signal. 2019, 30, 1062–1082. [Google Scholar] [CrossRef]
- Arias, I.M. Multidrug resistance genes, p-glycoprotein and the liver. Hepatology 1990, 12, 159–165. [Google Scholar] [CrossRef]
- Chen, F.W.; Gordon, R.E.; Ioannou, Y.A. NPC1 late endosomes contain elevated levels of non-esterified (‘free’) fatty acids and an abnormally glycosylated form of the NPC2 protein. Biochem. J. 2005, 390, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Geisslinger, F.; Müller, M.; Vollmar, A.M.; Bartel, K. Targeting Lysosomes in Cancer as Promising Strategy to Overcome Chemoresistance—A Mini Review. Front. Oncol. 2020, 10, 1156. [Google Scholar] [CrossRef] [PubMed]
- Tharkeshwar, A.K.; Demedts, D.; Annaert, W. Superparamagnetic Nanoparticles for Lysosome Isolation to Identify Spatial Alterations in Lysosomal Protein and Lipid Composition. STAR Protoc. 2020, 1, 100122. [Google Scholar] [CrossRef] [PubMed]






| Biochemical Data | Ultrastructural Data | |
|---|---|---|
| Enrichment of endolysosomes in F1? | Yes (indicated by LAMP-2, cathepsin D, and acid phosphatase). | Similar ultrastructure of the negatively stained F1 compared to the anti-LAMP-2 immunopurified vesicles. |
| Presence of Pgp in endolysosomes? | Possibly; indicated by (1) co-distribution of LAMP-2 (endolysosomal marker) and Pgp in F1 (however, by Western blot, a co-localization of Pgp and LAMP-2 cannot be determined but is not excluded); (2) detection of Pgp in the membrane fraction of F1; (3) Co-IP with LAMP-2 suggests that Pgp is present in the same vesicles but differentiation between localization at the limiting membrane and membranes of intraluminal vesicles (ILVs) is not possible. | Maybe in a subpopulation of small lysosomes (primary lysosomes?) as indicated by the ultrastructure of anti-LAMP-2 and anti-Pgp immunopurified vesicles; however, these small vesicles may also represent transport vesicles, intraluminal vesicles (ILVs) or early endosomes. |
| Expression of Pgp in the limiting membrane of endolysosomes? | Cannot be determined biochemically, since the membrane fraction of F1 will contain both the limiting membrane of endolysosomes and membranes of ILVs, and, as indicated by EEA1, the limiting membranes of early endosomes; however, the functional data with rhodamine 123 and tariquidar may indicate that Pgp is (weakly) active in organelles such as early endosomes and endolysosomes. | No clear evidence from negative and positive staining of anti-LAMP-2 and anti-Pgp immunopurified vesicles; endolysosomes are only present in the LAMP-2 positive fraction. In situ localization of Pgp and LAMP-2 in rat liver sections indicate that some endolysosomes may express Pgp in their limiting membrane, but—based on the ultrastructural data from F1—this seems to be an exception; in contrast, LAMP-2 is predominantly expressed at the limiting membrane of endolysosomes. |
| Contamination of F1 with early endosomes (EEA1)? | Yes | Yes, as indicated by the small vesicles in negative stained F1 fraction. |
| Contamination of F1 with canalicular plasma membranes (ABCB4)? | No | Vesiculated fragments of cell membranes cannot be excluded. |
| Contamination of F1 with ER (calnexin)? | Yes | A ribosome connected network was not visible in F1. |
| Contamination of F1 with mitochondria (VDAC)? | No | Mitochondria were not seen. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gericke, B.; Wienböker, I.; Brandes, G.; Löscher, W. Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver. Cells 2022, 11, 1556. https://doi.org/10.3390/cells11091556
Gericke B, Wienböker I, Brandes G, Löscher W. Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver. Cells. 2022; 11(9):1556. https://doi.org/10.3390/cells11091556
Chicago/Turabian StyleGericke, Birthe, Inka Wienböker, Gudrun Brandes, and Wolfgang Löscher. 2022. "Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver" Cells 11, no. 9: 1556. https://doi.org/10.3390/cells11091556
APA StyleGericke, B., Wienböker, I., Brandes, G., & Löscher, W. (2022). Is P-Glycoprotein Functionally Expressed in the Limiting Membrane of Endolysosomes? A Biochemical and Ultrastructural Study in the Rat Liver. Cells, 11(9), 1556. https://doi.org/10.3390/cells11091556







