Intestinal Microbiota Remodeling Protects Mice from Western Diet-Induced Brain Inflammation and Cognitive Decline
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice Maintenance and Treatment Regimens
2.2. Gene Expression Profiling
2.3. Western Blot Analysis
2.4. Biochemical Analysis
2.5. Electrophysiological Recording for Measuring Long-Term Potentiation (LTP)
2.6. Brain Lipidomics
2.7. Gut Microbiota Analysis Using 16S rRNA Gene Sequencing
2.8. Untargeted Metabolomics Profile
2.9. Bile Acid Quantification
2.10. Bioinformatics and Statistical Analysis
3. Results
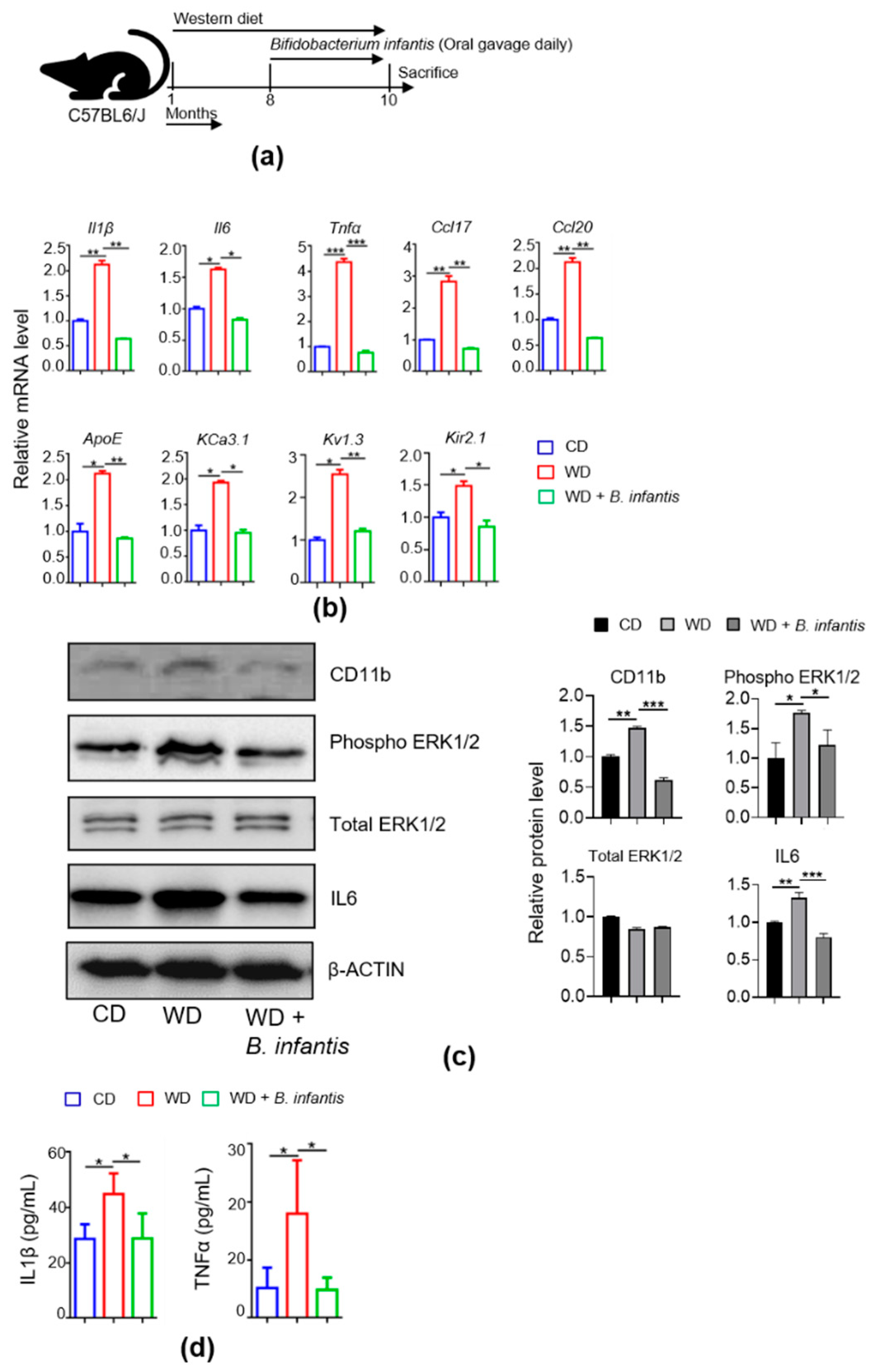
3.1. B. infantis Reduces diet-Induced Brain Inflammation
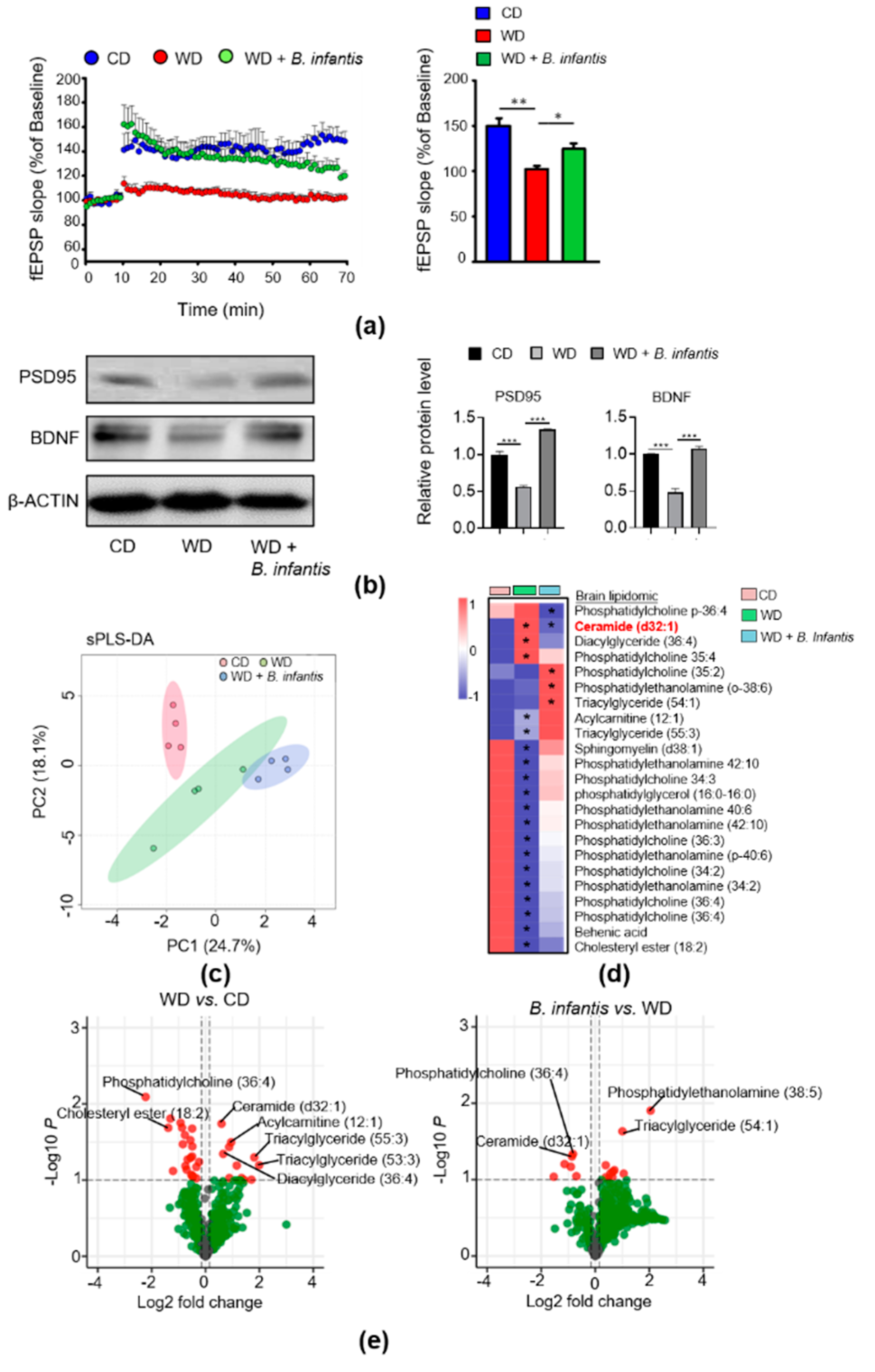
3.2. B. infantis Improves Neuroplasticity in WD-Fed Mice
3.3. B. infantis Supplementation Changes Brain Lipidomic Profiles in WD-Fed Mice
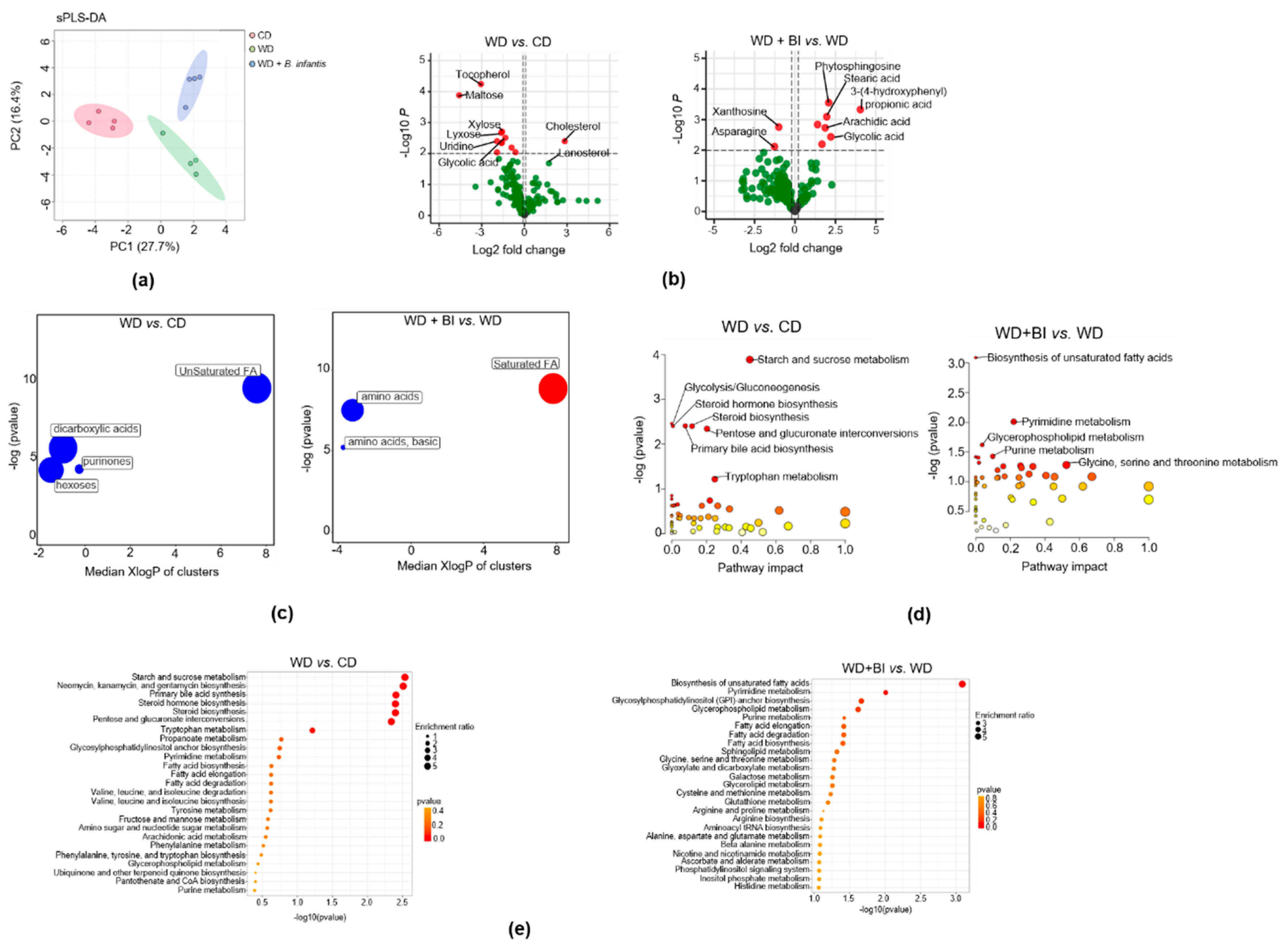
3.4. B. infantis Supplementation Modulates Cecal Metabolome
3.5. B. infantis Supplementation Alters the Gut Microbiota of WD-Fed Mice
3.6. Microbiome Depletion by Antibiotics Reduce Brain Inflammation and Increase BA Receptor Signaling
3.7. B. infantis Supplementation Enhances BA Receptor Signaling
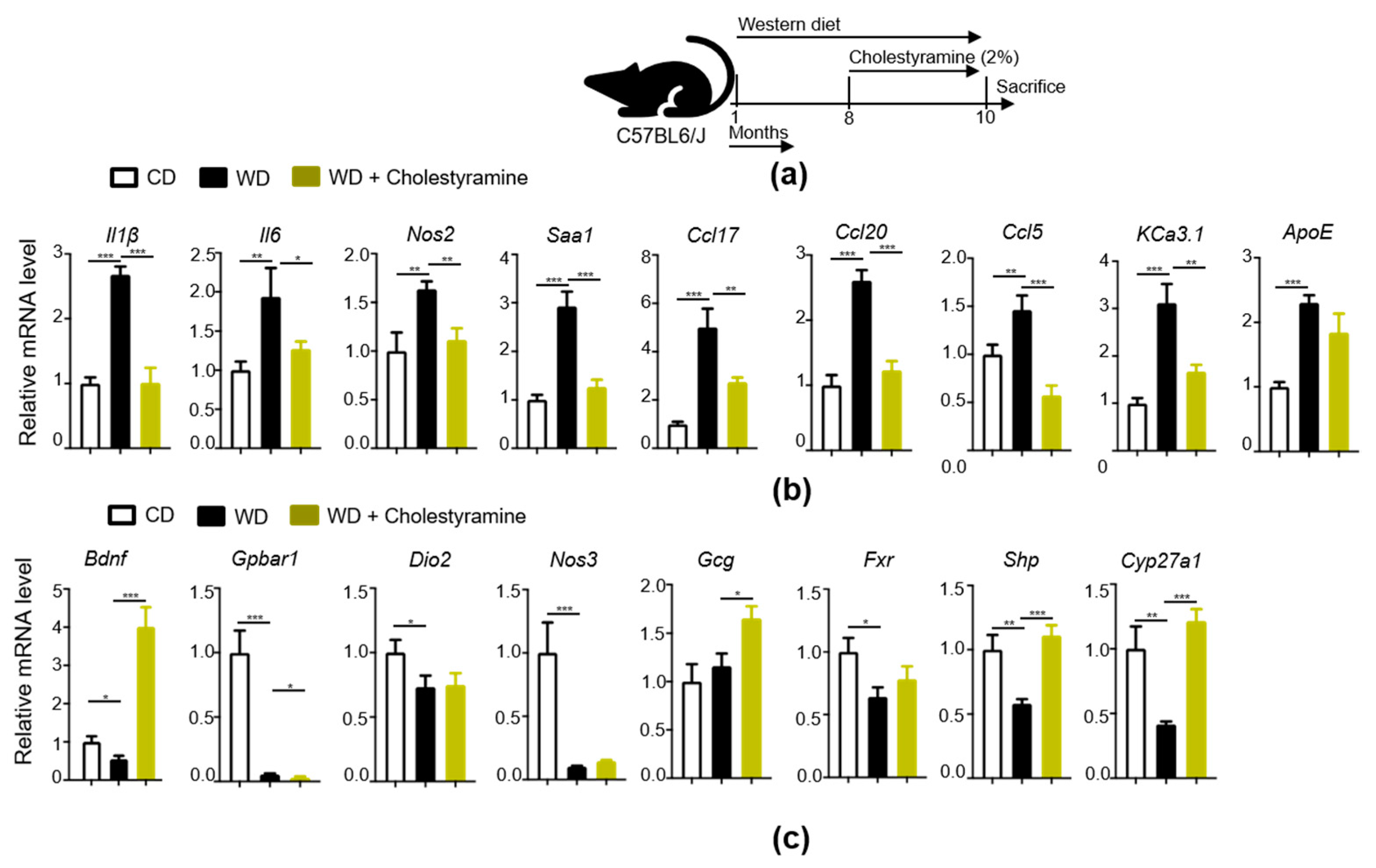
3.8. Bile Acid Sequestrant, Cholestyramine, Reduces WD-Indued Brain Inflammation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noble, E.E.; Hsu, T.M.; Kanoski, S.E. Gut to Brain Dysbiosis: Mechanisms Linking Western Diet Consumption, the Microbiome, and Cognitive Impairment. Front. Behav. Neurosci. 2017, 11, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jena, P.K.; Sheng, L.; Di Lucente, J.; Jin, L.W.; Maezawa, I.; Wan, Y.Y. Dysregulated bile acid synthesis and dysbiosis are implicated in Western diet-induced systemic inflammation, microglial activation, and reduced neuroplasticity. FASEB J. 2018, 32, 2866–2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanoski, S.E.; Davidson, T.L. Different patterns of memory impairments accompany short- and longer-term maintenance on a high-energy diet. J. Exp. Psychol. Anim. Behav. Processes 2010, 36, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.J.; Knight, N.S.; Cochlin, L.E.; McAleese, S.; Deacon, R.M.; Rawlins, J.N.; Clarke, K. Deterioration of physical performance and cognitive function in rats with short-term high-fat feeding. FASEB J. 2009, 23, 4353–4360. [Google Scholar] [CrossRef] [PubMed]
- Khazen, T.; Hatoum, O.A.; Ferreira, G.; Maroun, M. Acute exposure to a high-fat diet in juvenile male rats disrupts hippocampal-dependent memory and plasticity through glucocorticoids. Sci. Rep. 2019, 9, 12270. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Baym, C.L.; Monti, J.M.; Raine, L.B.; Drollette, E.S.; Scudder, M.R.; Moore, R.D.; Kramer, A.F.; Hillman, C.H.; Cohen, N.J. Central adiposity is negatively associated with hippocampal-dependent relational memory among overweight and obese children. J. Pediatr. 2015, 166, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Winocur, G.; Greenwood, C.E.; Piroli, G.G.; Grillo, C.A.; Reznikov, L.R.; Reagan, L.P.; McEwen, B.S. Memory impairment in obese Zucker rats: An investigation of cognitive function in an animal model of insulin resistance and obesity. Behav. Neurosci. 2005, 119, 1389–1395. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Jiang, Y.; Xu, K.; Cui, M.; Ye, W.; Zhao, G.; Jin, L.; Chen, X. The progress of gut microbiome research related to brain disorders. J. Neuroinflamm. 2020, 17, 25. [Google Scholar] [CrossRef] [Green Version]
- Arciszewski, M.B.; Sand, E.; Ekblad, E. Vasoactive intestinal peptide rescues cultured rat myenteric neurons from lipopolysaccharide induced cell death. Regul. Pept. 2008, 146, 218–223. [Google Scholar] [CrossRef]
- Yarandi, S.S.; Kulkarni, S.; Saha, M.; Sylvia, K.E.; Sears, C.L.; Pasricha, P.J. Intestinal Bacteria Maintain Adult Enteric Nervous System and Nitrergic Neurons via Toll-like Receptor 2-induced Neurogenesis in Mice. Gastroenterology 2020, 159, 200–213. [Google Scholar] [CrossRef]
- Hein, A.M.; Stasko, M.R.; Matousek, S.B.; Scott-McKean, J.J.; Maier, S.F.; Olschowka, J.A.; Costa, A.C.; O’Banion, M.K. Sustained hippocampal IL-1beta overexpression impairs contextual and spatial memory in transgenic mice. Brain Behav. Immun. 2010, 24, 243–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heumann, D.; Barras, C.; Severin, A.; Glauser, M.P.; Tomasz, A. Gram-positive cell walls stimulate synthesis of tumor necrosis factor alpha and interleukin-6 by human monocytes. Infect. Immun. 1994, 62, 2715–2721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza-Díaz, J.; Ruiz-Ojeda, F.J.; Vilchez-Padial, L.M.; Gil, A. Evidence of the Anti-Inflammatory Effects of Probiotics and Synbiotics in Intestinal Chronic Diseases. Nutrients 2017, 9, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divyashri, G.; Krishna, G.; Muralidhara; Prapulla, S.G. Probiotic attributes, antioxidant, anti-inflammatory and neuromodulatory effects of Enterococcus faecium CFR 3003: In vitro and in vivo evidence. J. Med. Microbiol. 2015, 64, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Akbari, E.; Asemi, Z.; Daneshvar Kakhaki, R.; Bahmani, F.; Kouchaki, E.; Tamtaji, O.R.; Hamidi, G.A.; Salami, M. Effect of Probiotic Supplementation on Cognitive Function and Metabolic Status in Alzheimer’s Disease: A Randomized, Double-Blind and Controlled Trial. Front. Aging Neurosci. 2016, 8, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldassarre, M.E.; Palladino, V.; Amoruso, A.; Pindinelli, S.; Mastromarino, P.; Fanelli, M.; Di Mauro, A.; Laforgia, N. Rationale of Probiotic Supplementation during Pregnancy and Neonatal Period. Nutrients 2018, 10, 1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.T.; Chen, S.; Jena, P.K.; Sheng, L.; Hu, Y.; Wan, Y.-J.Y. Probiotics Improve Gastrointestinal Function and Life Quality in Pregnancy. Nutrients 2021, 13, 3931. [Google Scholar] [CrossRef]
- Lawson, M.A.E.; O’Neill, I.J.; Kujawska, M.; Gowrinadh Javvadi, S.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020, 14, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Jena, P.K.; Sheng, L.; Nagar, N.; Wu, C.; Barile, D.; Mills, D.A.; Wan, Y.Y. Synbiotics Bifidobacterium infantis and milk oligosaccharides are effective in reversing cancer-prone nonalcoholic steatohepatitis using western diet-fed FXR knockout mouse models. J. Nutr. Biochem. 2018, 57, 246–254. [Google Scholar] [CrossRef]
- Minter, M.R.; Zhang, C.; Leone, V.; Ringus, D.L.; Zhang, X.; Oyler-Castrillo, P.; Musch, M.W.; Liao, F.; Ward, J.F.; Holtzman, D.M.; et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 2016, 6, 30028. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; Liu, H.X.; Kalanetra, K.M.; Mirsoian, A.; Murphy, W.J.; French, S.W.; Krishnan, V.V.; Mills, D.A.; Wan, Y.Y. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am. J. Pathol. 2017, 187, 1800–1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Elliott, C.T.; McGuinness, B.; Passmore, P.; Kehoe, P.G.; Hölscher, C.; McClean, P.L.; Graham, S.F.; Green, B.D. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer’s Disease. Metabolites 2017, 7, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, M.; McMillin, M.; Galindo, C.; Frampton, G.; Pae, H.Y.; DeMorrow, S. Bile acids permeabilize the blood brain barrier after bile duct ligation in rats via Rac1-dependent mechanisms. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2014, 46, 527–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baloni, P.; Funk, C.C.; Yan, J.; Yurkovich, J.T.; Kueider-Paisley, A.; Nho, K.; Heinken, A.; Jia, W.; Mahmoudiandehkordi, S.; Louie, G.; et al. Metabolic Network Analysis Reveals Altered Bile Acid Synthesis and Metabolism in Alzheimer’s Disease. Cell Rep. Med. 2020, 1, 100138. [Google Scholar] [CrossRef] [PubMed]
- Marksteiner, J.; Blasko, I.; Kemmler, G.; Koal, T.; Humpel, C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 2018, 14, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olazarán, J.; Gil-de-Gómez, L.; Rodríguez-Martín, A.; Valentí-Soler, M.; Frades-Payo, B.; Marín-Muñoz, J.; Antúnez, C.; Frank-García, A.; Acedo-Jiménez, C.; Morlán-Gracia, L.; et al. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer’s disease. J. Alzheimers Dis. 2015, 45, 1157–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maezawa, I.; Jenkins, D.P.; Jin, B.E.; Wulff, H. Microglial KCa3.1 Channels as a Potential Therapeutic Target for Alzheimer’s Disease. Int. J. Alzheimer’s Dis. 2012, 2012, 868972. [Google Scholar] [CrossRef] [Green Version]
- Ismail, I.T.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H.; Fiehn, O. Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers 2020, 13, 88. [Google Scholar] [CrossRef]
- Sheng, L.; Jena, P.K.; Liu, H.X.; Hu, Y.; Nagar, N.; Bronner, D.N.; Settles, M.L.; Bäumler, A.J.; Wan, Y.Y. Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J. 2018, 32, fj201800370R. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Lee, D.Y.; Lu, Y.; Moon, S.; Nikolau, B. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant. J. 2008, 53, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Jena, P.K.; Sheng, L.; Nguyen, M.; Di Lucente, J.; Hu, Y.; Li, Y.; Maezawa, I.; Jin, L.-W.; Wan, Y.-J.Y. Dysregulated bile acid receptor-mediated signaling and IL-17A induction are implicated in diet-associated hepatic health and cognitive function. Biomark. Res. 2020, 8, 59. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- García-Cañaveras, J.C.; Donato, M.T.; Castell, J.V.; Lahoz, A. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J. Lipid Res. 2012, 53, 2231–2241. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Sun, J.; Nan, G. The extracellular signal-regulated kinase 1/2 pathway in neurological diseases: A potential therapeutic target (Review). Int J. Mol. Med. 2017, 39, 1338–1346. [Google Scholar] [CrossRef] [Green Version]
- Castellanos, D.B.; Martín-Jiménez, C.A.; Rojas-Rodríguez, F.; Barreto, G.E.; González, J. Brain lipidomics as a rising field in neurodegenerative contexts: Perspectives with Machine Learning approaches. Front. Neuroendocrinol. 2021, 61, 100899. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.S.; Tiwari, N.K. Lipid Integration in Neurodegeneration: An Overview of Alzheimer’s Disease. Mol. Neurobiol. 2014, 50, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-j.; Li, G.-m.; Tang, W.-l.; Yin, M. Neuroprotective effects of stearic acid against toxicity of oxygen/glucose deprivation or glutamate on rat cortical or hippocampal slices. Acta Pharmacol. Sin. 2006, 27, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallis, T.P.; Venkatesh, B.G.; Narayana, V.K.; Kvaskoff, D.; Ho, A.; Sullivan, R.K.; Windels, F.; Sah, P.; Meunier, F.A. Saturated free fatty acids and association with memory formation. Nat. Commun. 2021, 12, 3443. [Google Scholar] [CrossRef] [PubMed]
- Bekinschtein, P.; Cammarota, M.; Katche, C.; Slipczuk, L.; Rossato, J.I.; Goldin, A.; Izquierdo, I.; Medina, J.H. BDNF is essential to promote persistence of long-term memory storage. Proc. Natl. Acad. Sci. USA 2008, 105, 2711–2716. [Google Scholar] [CrossRef] [Green Version]
- Jiao, S.S.; Shen, L.L.; Zhu, C.; Bu, X.L.; Liu, Y.H.; Liu, C.H.; Yao, X.Q.; Zhang, L.L.; Zhou, H.D.; Walker, D.G.; et al. Brain-derived neurotrophic factor protects against tau-related neurodegeneration of Alzheimer’s disease. Transl. Psychiatry 2016, 6, e907. [Google Scholar] [CrossRef]
- Hansen, S.N.; Ipsen, D.H.; Schou-Pedersen, A.M.; Lykkesfeldt, J.; Tveden-Nyborg, P. Long term Westernized diet leads to region-specific changes in brain signaling mechanisms. Neurosci. Lett. 2018, 676, 85–91. [Google Scholar] [CrossRef]
- Wu, A.; Ying, Z.; Gomez-Pinilla, F. The interplay between oxidative stress and brain-derived neurotrophic factor modulates the outcome of a saturated fat diet on synaptic plasticity and cognition. Eur. J. Neurosci. 2004, 19, 1699–1707. [Google Scholar] [CrossRef]
- Setayesh, T.; Mišík, M.; Langie, S.A.S.; Godschalk, R.; Waldherr, M.; Bauer, T.; Leitner, S.; Bichler, C.; Prager, G.; Krupitza, G.; et al. Impact of Weight Loss Strategies on Obesity-Induced DNA Damage. Mol. Nutr. Food Res. 2019, 63, e1900045. [Google Scholar] [CrossRef]
- Tian, P.; Zou, R.; Song, L.; Zhang, X.; Jiang, B.; Wang, G.; Lee, Y.K.; Zhao, J.; Zhang, H.; Chen, W. Ingestion of Bifidobacterium longum subspecies infantis strain CCFM687 regulated emotional behavior and the central BDNF pathway in chronic stress-induced depressive mice through reshaping the gut microbiota. Food Funct. 2019, 10, 7588–7598. [Google Scholar] [CrossRef]
- El-Husseini Ael, D.; Schnell, E.; Dakoji, S.; Sweeney, N.; Zhou, Q.; Prange, O.; Gauthier-Campbell, C.; Aguilera-Moreno, A.; Nicoll, R.A.; Bredt, D.S. Synaptic strength regulated by palmitate cycling on PSD-95. Cell 2002, 108, 849–863. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yoon, G.; Song, J.; Jo, J. Exendin-4 improves long-term potentiation and neuronal dendritic growth in vivo and in vitro obesity condition. Sci. Rep. 2021, 11, 8326. [Google Scholar] [CrossRef]
- Power, M.C.; Rawlings, A.; Sharrett, A.R.; Bandeen-Roche, K.; Coresh, J.; Ballantyne, C.M.; Pokharel, Y.; Michos, E.D.; Penman, A.; Alonso, A.; et al. Association of midlife lipids with 20-year cognitive change: A cohort study. Alzheimers Dement. 2018, 14, 167–177. [Google Scholar] [CrossRef]
- Tong, M.; de la Monte, S.M. Mechanisms of ceramide-mediated neurodegeneration. J. Alzheimers Dis 2009, 16, 705–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alessenko, A.V.; Bugrova, A.E.; Dudnik, L.B. Connection of lipid peroxide oxidation with the sphingomyelin pathway in the development of Alzheimer’s disease. Biochem. Soc. Trans. 2004, 32, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Filippov, V.; Song, M.A.; Zhang, K.; Vinters, H.V.; Tung, S.; Kirsch, W.M.; Yang, J.; Duerksen-Hughes, P.J. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J. Alzheimers Dis 2012, 29, 537–547. [Google Scholar] [CrossRef] [Green Version]
- Lyn-Cook, L.E., Jr.; Lawton, M.; Tong, M.; Silbermann, E.; Longato, L.; Jiao, P.; Mark, P.; Wands, J.R.; Xu, H.; de la Monte, S.M. Hepatic ceramide may mediate brain insulin resistance and neurodegeneration in type 2 diabetes and non-alcoholic steatohepatitis. J. Alzheimers Dis. 2009, 16, 715–729. [Google Scholar] [CrossRef] [Green Version]
- Holland, W.L.; Brozinick, J.T.; Wang, L.P.; Hawkins, E.D.; Sargent, K.M.; Liu, Y.; Narra, K.; Hoehn, K.L.; Knotts, T.A.; Siesky, A.; et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007, 5, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Le Roy, T.; Lécuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barceló, J.; Huby, T.; Guerin, M.; et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-S.; Kim, Y.; Choi, H.; Kim, W.; Park, S.; Lee, D.; Kim, D.K.; Kim, H.J.; Choi, H.; Hyun, D.-W.; et al. Transfer of a healthy microbiota reduces amyloid and tau pathology in an Alzheimer’s disease animal model. Gut 2020, 69, 283–294. [Google Scholar] [CrossRef]
- van Vliet, P. Cholesterol and late-life cognitive decline. J. Alzheimers Dis 2012, 30, S147–S162. [Google Scholar] [CrossRef] [PubMed]
- Djelti, F.; Braudeau, J.; Hudry, E.; Dhenain, M.; Varin, J.; Bièche, I.; Marquer, C.; Chali, F.; Ayciriex, S.; Auzeil, N.; et al. CYP46A1 inhibition, brain cholesterol accumulation and neurodegeneration pave the way for Alzheimer’s disease. Brain 2015, 138, 2383–2398. [Google Scholar] [CrossRef] [Green Version]
- Fu, B.Y.; Ma, S.L.; Tang, N.L.; Tam, C.W.; Lui, V.W.; Chiu, H.F.; Lam, L.C. Cholesterol 24-hydroxylase (CYP46A1) polymorphisms are associated with faster cognitive deterioration in Chinese older persons: A two-year follow up study. Int. J. Geriatr. Psychiatry 2009, 24, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Pikuleva, I.A.; Cartier, N. Cholesterol Hydroxylating Cytochrome P450 46A1: From Mechanisms of Action to Clinical Applications. Front. Aging Neurosci. 2021, 13, 696778. [Google Scholar] [CrossRef]
- Houten, S.M.; Watanabe, M.; Auwerx, J. Endocrine functions of bile acids. EMBO J. 2006, 25, 1419–1425. [Google Scholar] [CrossRef]
- Griffiths, W.J.; Sjövall, J. Bile acids: Analysis in biological fluids and tissues. J. Lipid Res. 2010, 51, 23–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MahmoudianDehkordi, S.; Arnold, M.; Nho, K.; Ahmad, S.; Jia, W.; Xie, G.; Louie, G.; Kueider-Paisley, A.; Moseley, M.A.; Thompson, J.W.; et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimers Dement. 2019, 15, 76–92. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Lee, B.H. Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. Publ. Protein Soc. 2018, 27, 1742–1754. [Google Scholar] [CrossRef]
- Mertens, K.L.; Kalsbeek, A.; Soeters, M.R.; Eggink, H.M. Bile Acid Signaling Pathways from the Enterohepatic Circulation to the Central Nervous System. Front. Neurosci. 2017, 11, 617. [Google Scholar] [CrossRef] [Green Version]
- Schubring, S.R.; Fleischer, W.; Lin, J.S.; Haas, H.L.; Sergeeva, O.A. The bile steroid chenodeoxycholate is a potent antagonist at NMDA and GABA(A) receptors. Neurosci. Lett. 2012, 506, 322–326. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; McNeil, K.; Chau, T.Q.; Yu, S.; Kiuru, M.; Fung, M.A.; Hwang, S.T.; Wan, Y.-J.Y. Long-term Western diet intake leads to dysregulated bile acid signaling and dermatitis with Th2 and Th17 pathway features in mice. J. Dermatol. Sci. 2019, 95, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMillin, M.; Frampton, G.; Quinn, M.; Ashfaq, S.; de los Santos, M., 3rd; Grant, S.; DeMorrow, S. Bile Acid Signaling Is Involved in the Neurological Decline in a Murine Model of Acute Liver Failure. Am. J. Pathol 2016, 186, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Rust, C.; Sauter, G.H.; Oswald, M.; Büttner, J.; Kullak-Ublick, G.A.; Paumgartner, G.; Beuers, U. Effect of cholestyramine on bile acid pattern and synthesis during administration of ursodeoxycholic acid in man. Eur. J. Clin. Invest. 2000, 30, 135–139. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jena, P.K.; Setayesh, T.; Sheng, L.; Di Lucente, J.; Jin, L.W.; Wan, Y.-J.Y. Intestinal Microbiota Remodeling Protects Mice from Western Diet-Induced Brain Inflammation and Cognitive Decline. Cells 2022, 11, 504. https://doi.org/10.3390/cells11030504
Jena PK, Setayesh T, Sheng L, Di Lucente J, Jin LW, Wan Y-JY. Intestinal Microbiota Remodeling Protects Mice from Western Diet-Induced Brain Inflammation and Cognitive Decline. Cells. 2022; 11(3):504. https://doi.org/10.3390/cells11030504
Chicago/Turabian StyleJena, Prasant Kumar, Tahereh Setayesh, Lili Sheng, Jacopo Di Lucente, Lee Way Jin, and Yu-Jui Yvonne Wan. 2022. "Intestinal Microbiota Remodeling Protects Mice from Western Diet-Induced Brain Inflammation and Cognitive Decline" Cells 11, no. 3: 504. https://doi.org/10.3390/cells11030504






